MCSC-Net: COVID-19 detection using deep-Q-neural network classification with RFNN-based hybrid whale optimization
Abstract
BACKGROUND:
COVID-19 is the most dangerous virus, and its accurate diagnosis saves lives and slows its spread. However, COVID-19 diagnosis takes time and requires trained professionals. Therefore, developing a deep learning (DL) model on low-radiated imaging modalities like chest X-rays (CXRs) is needed.
OBJECTIVE:
The existing DL models failed to diagnose COVID-19 and other lung diseases accurately. This study implements a multi-class CXR segmentation and classification network (MCSC-Net) to detect COVID-19 using CXR images.
METHODS:
Initially, a hybrid median bilateral filter (HMBF) is applied to CXR images to reduce image noise and enhance the COVID-19 infected regions. Then, a skip connection-based residual network-50 (SC-ResNet50) is used to segment (localize) COVID-19 regions. The features from CXRs are further extracted using a robust feature neural network (RFNN). Since the initial features contain joint COVID-19, normal, pneumonia bacterial, and viral properties, the conventional methods fail to separate the class of each disease-based feature. To extract the distinct features of each class, RFNN includes a disease-specific feature separate attention mechanism (DSFSAM). Furthermore, the hunting nature of the Hybrid whale optimization algorithm (HWOA) is used to select the best features in each class. Finally, the deep-Q-neural network (DQNN) classifies CXRs into multiple disease classes.
RESULTS:
The proposed MCSC-Net shows the enhanced accuracy of 99.09% for 2-class, 99.16% for 3-class, and 99.25% for 4-class classification of CXR images compared to other state-of-art approaches.
CONCLUSION:
The proposed MCSC-Net enables to conduct multi-class segmentation and classification tasks applying to CXR images with high accuracy. Thus, together with gold-standard clinical and laboratory tests, this new method is promising to be used in future clinical practice to evaluate patients.
1Introduction
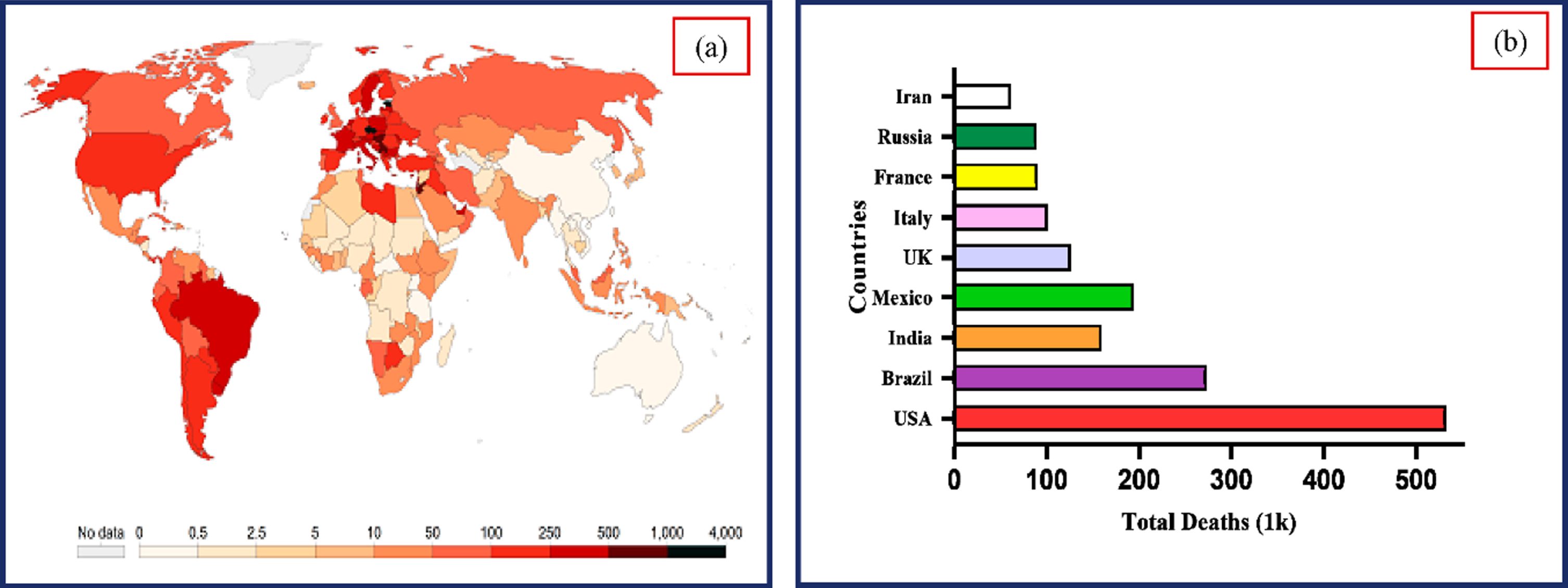
According to reports, the COVID-19 virus causes lung damage and rapidly mutates before the patient receives any diagnosis-specific medicine. The situation gets more dangerous when the symptoms resemble the ordinary flu, as they did in Southeast Asia and Central Asia. The World Health Organization (WHO) declared COVID-19, a contagious illness brought back by the SARS virus, a global pandemic in March 2020. As of December 2, 2022, the massive COVID-19 epidemic has spread to 640.39 million individuals and caused more than 6.61 million death incidents globally [49]. As indicated in Fig. 1, the infected cases and fatality rate are growing significantly. Early diagnosis of COVID-19 is critical to restrict the wide spread of the virus and offer treatment to avoid consequences [30]. It is challenging to recognize and control the pandemic due to the increase in COVID-19 incidents globally and the limits of the currently used diagnostic tools.
Fig. 1
(a) Worldwide COVID-19 incidence map (b). Total COVID-19 deaths reported in various nations. (Source: Center for Systems Science and Engineering at Johns Hopkins University, Baltimore, MD, USA).
Globally engaged researchers [20] are expediting the development of vaccines and treatments and searching for new diagnostic methods. Blood testing, virus tests, and medical imaging [13] are standard diagnostic procedures in the United States. Blood tests can identify antibodies against the coronavirus-2 (SARS-CoV-2) that causes severe acute respiratory syndrome (SARS). The antigens of SARS-CoV-2 are detected utilizing viral assays on samples taken from the respiratory tract by a rapid antigens diagnostic test kits (RDT) [35]. It is a quick test that may provide results in as little as 30 minutes. The efficiency of this RDT test kits depends on the sample’s quality and the period at which the sickness first manifests itself. Furthermore, since the test does not discriminate COVID-19 from viral infections, it can potentially provide false-positive findings. The reverse transcription-polymerase chain reaction (RT-PCR) is the gold-standard technology for first-line screening [12]. However, a thorough investigation has shown that the test results’ sensitivity varies between 50 and 62 percent. Consequently, repeated RT-PCR tests are done during a 14-day observation period to guarantee that the test result is accurate for diagnosis [1] as described above. Patients could become frustrated because there aren’t many RT-PCR test kits available in different nations [26], which could be costly for healthcare organizations. Medical imaging technique like chest computer tomography (CT) is frequently used to diagnose pneumonia due to COVID-19 [2, 16, 51]. Computer tomography (CT) is more sensitive for early pneumonic change, illness development, and alternative diagnoses; in this situation, intravenous contrast material injection is necessary for diagnosing pulmonary embolism. Imaging technologies alone are insufficient to diagnose COVID-19 pneumonia, despite recent advancements in diagnostic techniques [36].
Chest Computer Tomography (Chest CT), and Magnetic Resonance Imaging (MRI) are more radiation diagnostic tools not recommended as per the current guidelines. The most distinctive feature of the COVID-19 sickness was the bilateral ground-glass opacities (GGO) [4], which may or may not be accompanied by consolidations in lungs. Pulmonary embolism and a block in an artery caused by blood clots may be more common in COVID-19 individuals. The situation becomes more problematic when it comes to CT scan examination due to the use of contrast [3, 5, 37]. Chest radiography scans are essential for detecting and treating COVID-19 as early as feasible because the virus affects the respiratory system.
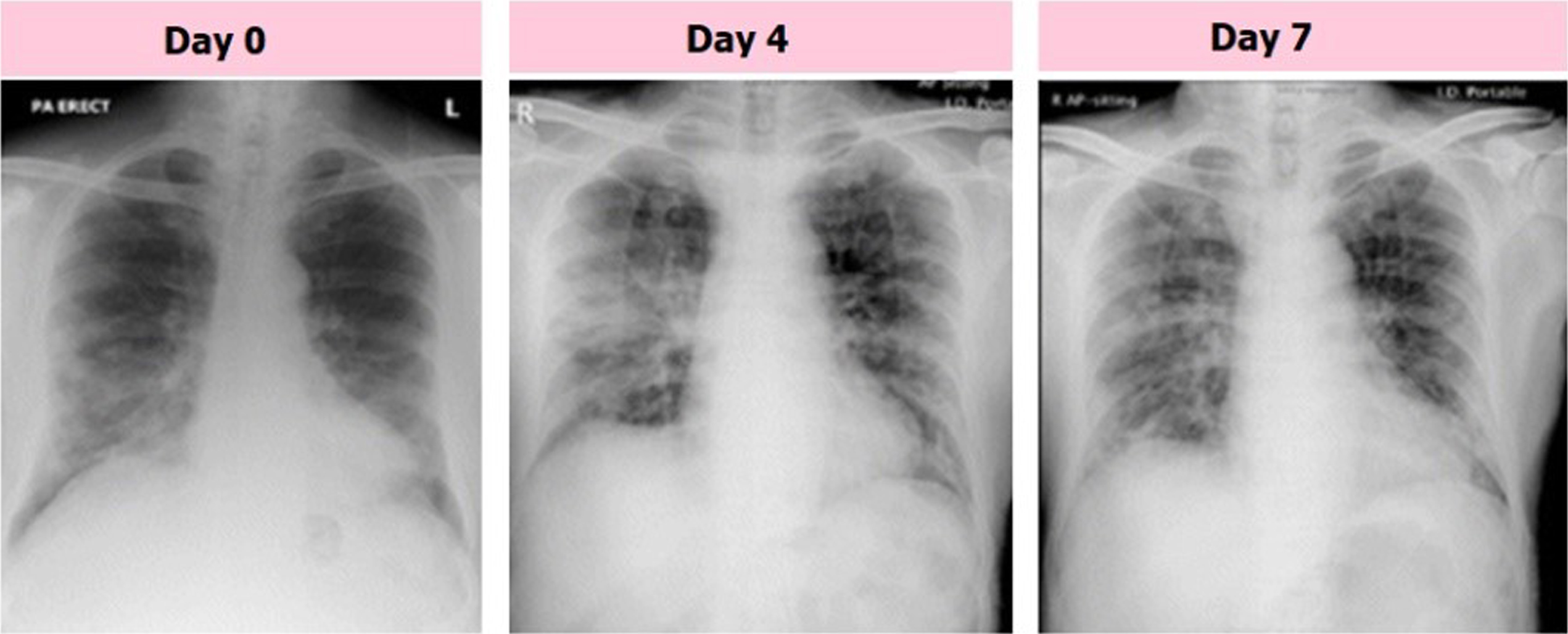
The chest radiographs of a senior citizen from Wuhan, China, who traveled to Hong Kong, China, for medical care are shown in Fig. 2. Fresh consolidative alterations are now seen in the right midzone perimeter and perihilar area of the right lower zone, where the consolidation that started on day 0 has persisted into day 4 of the zone. The day 7 footage’s midzone adjustment, which features a high GGO area, is an improvement over the day before. Consequently, CXRs has been employed as a primary imaging diagnosis tool in many nations in on-going pandemic. With the use of radiology scans, it is possible to determine the status of the lungs and phases of disease that are taking place. The CXRs of COVID-19 individuals were subjected to various anomalies, which radiologists noted. CXR is a commonly accessible scan for chest with no patient preparation required to diagnose disease with instantaneous result. A CT scan may be used to perform tasks such as patient triage, determining which therapies are most critical for patients, and determining how best to utilize available medical resources. CT is more sensitive to disease progression, early pneumonic alteration, and other diagnoses. An intravenous contrast medium is needed to identify pulmonary embolism. Imaging technologies alone are insufficient to diagnose COVID-19 pneumonia, despite recent advancements in diagnostic techniques. Imaging should be used with lab and clinical testing. COVID-19 chest CTs show bilateral, peripheral, and basal GGOs with or without consolidation. CT screening could not reveal GGO characteristics. Due to this, CXR screening is recommended.
Fig. 2
Samples of COVID-19 affected CXR images with GGO consolidation.
CXRs were employed in several studies [31, 39, 48] for COVID-19 diagnosis and classification, and the results were favorable compared to CT-based COVID-19 diagnosis [6, 14], which uses radiological images such as CXR images. The CXR has several advantages over CT, including rapid data acquisition, reduced ionizing radiation, portability, and more availability in intensive care units (ICU). These benefits make CXR a useful tool for radiologists. The most significant contributions made by this work are listed below:
• This work proposed multi-class CXR segmentation and classification network (MCSC-Net) for detecting diseases presented in the CXR images along with COVID-19.
• The MCSC-Net contains multiple stages of operation, such as HMBF for preprocessing, SC-ResNet50 for segmentation, RFNN with DSFSAM for feature extraction, optimal feature selection by HWOA, and DQNN classification.
• The MCSC-Net model is also implemented as the 2-class, 3-class, and 4-class model for both segmentation and classification operations, which helps to check the reliability of the overall system.
• The MCSC-Net perfectly separated the disease-specific and disease-dependent features of COVID-19, bacterial and viral pneumonia, and normal classes, which resulted in superior performance.
• The outcome demonstrates that, compared to traditional methods, the suggested MCSC-Net produced higher preprocessing, segmentation, and classification performance.
The article sections continues with the following structure: Section 2 discusses the literature survey and their limitations; Section 3 provides working of the proposed MCSC-Net with multiple stages, Section 4 analyzing the results with performance comparison using various existing works, and the conclusions along with possible future challenges are presented in Section 5.
2Literature survey
This section thoroughly analyzes machine, deep, and transfer learning-based strategies for diagnosing COVID-19. This literature survey focuses exclusively on feature extraction, segmentation, and classification techniques. Additionally, this literature concentrated on feature selection methods utilizing bio-optimization algorithms.
2.1Survey on CXR segmentation
The COVID-19 region can be localized using the CXR image segmentation techniques described in this section, which also aids in classifying COVID-19. L. O. Teixeira et al. [27] developed CNN architectures for segmentation and classification. The UNet is utilized to perform segmentation, and the VGG16, ResNet50V2, and InceptionV3 are used as classifiers. However, there are computational challenges with this approach. L. Zhang et al. [28] implemented a modified U-Net model with dual encoder fusion, namely DEFU-Net, for CXR image segmentation with accuracy of 98.04%. The computational complexity increases as the network’s depth increase due to the densely connected recurrent CNN blocks. The FractalCovNet, the combination of fractal blocks and the U-Net, is developed by H. Munusamy et al. [23] for the localization of the lesion regions in the CXR and CT images. Further, the FractalCovNet with transfer learning approach is presented to classify the CXR images and suffers from the complexities. The pneumothorax boundary segmentation is perceived effectively by the Deep Signed Distance Map (DeepSDM) proposed by Y. Wang et al. [50]. The classification of CXRs using DeepSDM resulted in missed diagnosis and lower accuracy. S.Tabik et al. [40] presented a COVID-SDNet to improve detection accuracy by combining segmentation, data transformation, and augmentation. Class inherent-based methods are used to differentiate the classification capability of the classifiers. However, these inherence approaches yield lower classification accuracy. S. Motamed et al. [41] implemented a transfer learning (TL) approach for segmenting the lungs in the CXRs and presented randomized GAN (RANDGAN) to identify the images of the unknown class in CXRs. But these methods exhibit low accuracy with high computational complexity.
A variational auto-encoder (VAE) is introduced in the encoder-decoder of UNet to perform the lung lobes segmentation in CXR images by F. Cao et al. [21]. The extracted features are recognized highly due to the attention mechanism. The opacities are affecting the accuracy of segmenting in the majority of the lung regions. M. Kim et al. [32] developed a deep-learning neural network with a self-attention (SADNN) concept to perform automatic lung segmentation in CXR images by modifying the U-Net architecture. However, the segmentation performance depends on the dataset, and this method fails in achieving the segmentation if the dataset contains deformed shapes or lesions in the lungs. The fast and efficient multi-task DL (MTDL) approaches with COVID_MTNet is developed to diagnose COVID-19 in CT and CXRs by M. Z. Alom et al. [33]. These approaches utilize the NABLA-N model to perform the segmentation of both images. But the classification accuracy of the CXR dataset is as low as 84.67%. A Structure Correcting (SA) Network based on the GAN is presented to perform segmentation of lungs in CXR images [22]. This SA network has a similar function to the general adversarial network (GAN). Attention-based-UNet is the backbone network for SAGAN. Three classes are employed to categorize the CXR images using a fully connected network. But the classification accuracy and the dice scores could be higher. The detection of COVID-19 in CXRs using deep TL networks such as ResNet, InceptionV, and the combination of these networks are developed by A. Narin et al. [7]. Three different datasets were analyzed to know the capacity of the five pre-trained models: transfer-learned CNNs (TL-CNNs). Still, the detection accuracy is 96.1% on two class classification datasets. A novel DenseCapsNet is developed by fusing the Dense and CapsNet to classify the CXR images [24]. Before classification, the CXR images are segmented for the lung lobes by the ultimately selected TernausNet. The 4-class classification accuracy is low in the fused network.
2.2Survey on optimization with classification methods
The many feature selection techniques that use evolutionary and natural selection-based features are described in detail in this section. Compared to deep learning feature selections, these strategies produced better features. The classification of different classes in CXR images is proposed in two stages by the advanced squirrel search optimization (ASSOA). The ResNet-50 in the first stage extracts the features, and the CNN learns the features. In the multi-layer perceptron (MLP) neural network, the ASSOA chooses the features and optimizes the weights. The presented algorithm is computationally complex [17]. The ability of imbalanced exploitation and poor diversity with local optima is the significant problems associated with a meta-heuristic slime mold algorithm (SMA). The quasi-reflection with SMA (QRSMA) is developed to improve the performance of the SMA [42]. This combination increases the segmentation problems in CXR images. However, this method fails at the multiple objective optimization problems. The QRSMA performs the classification of CXR images with CNN. The projected algorithm is developed by introducing the concept of population reduction in modified whale optimization (mWOAPR), which performs CXR image segmentation to detect COVID-19 [43]. In classifying CXRs, the population reduction method with a support vector machine (SVM) produces higher accuracy. P. Bhowal et al. [38] presented VGG-16, Xception-Net, and InceptionV3-Net to detect COVID-19 by feature extractions in the CXR images. A two-tier process selects the features in CXRs. With a lower accuracy of 93.33%, the InceptionV3-Net extracts features. A. T. Sahlol et al. [8] used fractional-order, and marine predators swarm optimization techniques (FO-MPA) to identify COVID-19 CXR image features. The CNN classifies the selected features. The Aquila is a swarm optimization algorithm that selects the best features from the features extracted by the MobileNet-V3 in CXRs [34]. This method effectively reduces the dimensionality of the image representation while improving classification accuracy.
To find COVID-19 present in the CXR images, an optimized CNN is developed by optimizing the CNN with the grey wolf optimization (GWO) technique. The OptCoNet is the network designed to identify the CXRs for COVID-19 [46]. The GWO and the hyperparameters select the optimized features to train the CNN are optimized by the same GWO. However, the classification accuracy could be higher on the dataset used by this study. The XGBoost utilizes particle swarm optimization (PSO) to extract the deep features in the X-ray images to predict COVID-19 in CXRs. Deep feature extraction uses VGG-19, InceptionV3, and ResNet-50 DL networks [15]. The PSO-optimized XGBoost selects the best deep-optimized features. However, these methods of deep feature extraction are computationally complex. To detect COVID-19, the computer tomographic image (CT) features are retrieved and used with the CNN. The retrieved features are selected by the genetic algorithm (GA), and four different classifiers are presented to classify the best features [18]. E.-S. M. El-Kenawy et al. [19] presented a feature selection and classification algorithm to classify CT images for COVID-19. The AlexNet-CNN model extracts the features, and the guided GWO algorithm selects optimal features. Furthermore, a voting classifier is developed to classify the selected features and diagnose COVID-19. A. Al-zubidi et al. [9] proposed fuzzy c-mean (FCM) and back propagation-CNN (BP-CNN) to classify COVID-19. The features extracted affect the classification performance of the FCM and BPA. The information gain (IG) mechanism is introduced to improve the FCM and BP-CNN classification accuracy. T. Goel et al. [47] implemented a GAN network to generate sufficient images in the dataset. WOA is used to tune the hyperparameters of GAN to enhance classification performance. A CNN model optimized to detect COVID-19 from CXR images was given by S. Pathan et al. [44]. The GWO, whale, and BAT algorithms carry out the optimization of the CNN model. To optimize the classification results, the hyperparameters are modified automatically. In [10], SegNet, U-Net, Hybrid CCN, and their optimization using grey wolf optimization (GWO) are provided for semantic segmentation of CXR images. However, because of a lack of ground truth images, networks may need to be able to localize COVID-19 infection. In [11], a CXGNet is present to detect the COVID-19, but the classification accuracy is low. One major drawback of earlier research utilizing COVID-19 detection from CXR and CT images is their low detection accuracy. However, prior studies did not differentiate the distinct characteristics of the several classes of CXRs. This study aims to localize the specific features in the CXR classes and make the DL network suitable for the multi-class classification of diseases in the CXR images. The classification accuracy can be improved with the proposed network of this work due to the Disease-Specific Feature Separate Attention Mechanism (DSFSAM) to extract the separate features of each class.
3Proposed method
Methods including Transfer learning, Deep learning, Q-learning, and Bio-optimization are used to generate the proposed model described in this section. The block diagram of the proposed MCSC-Net for multi-class classification of CXR images is shown in Fig. 3.
Fig. 3
Block diagram of proposed MCSC-Net.
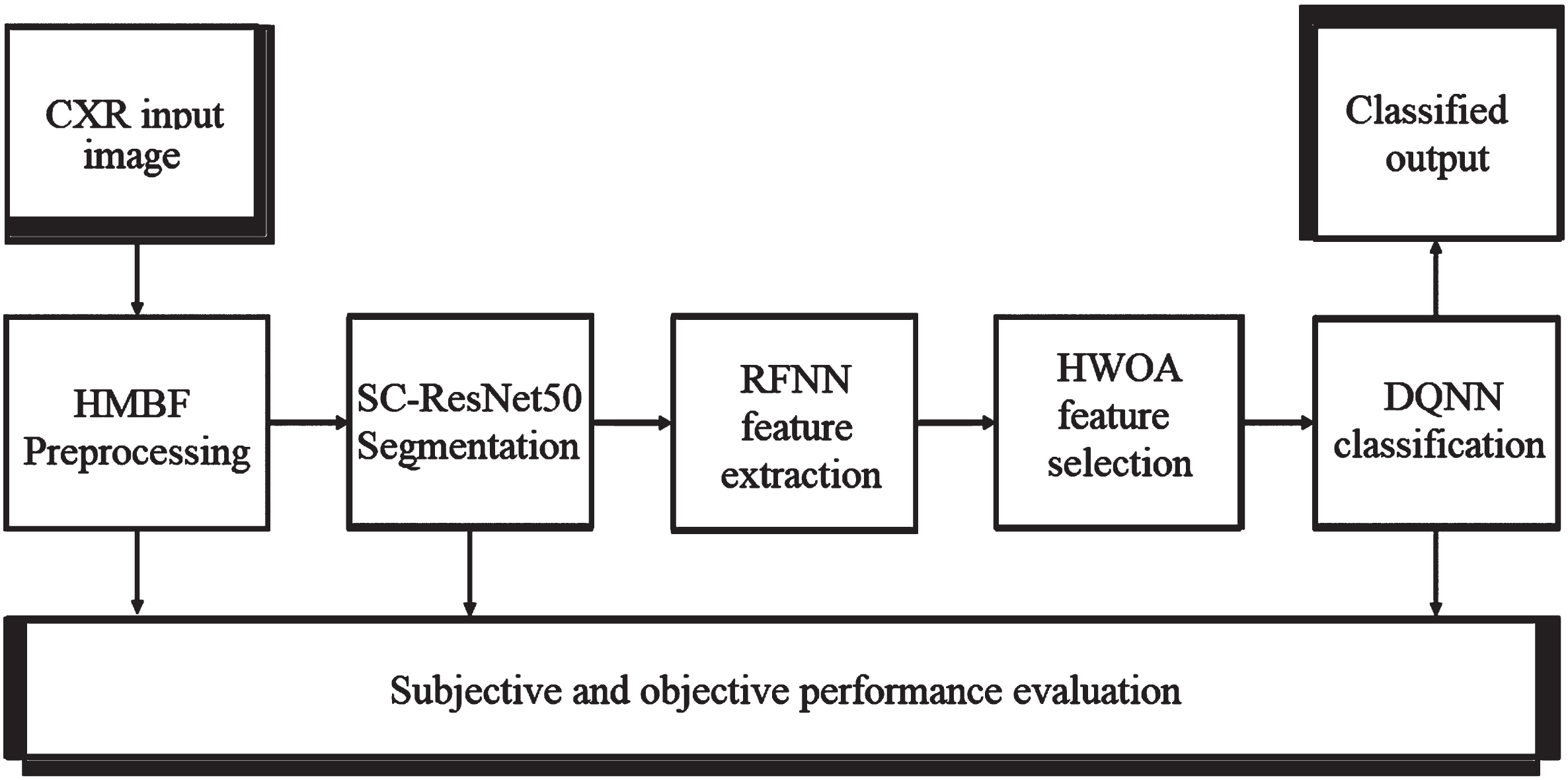
The Hybrid Median Bilateral Filter (HMBF) is first applied to CXR images to remove various types of noise, including random, Gaussian, salt, and pepper noise. Further, it is used to improve the COVID-19 area by enhancing the CXR images. Once this has been accomplished, the segmentation process is carried out using transfer learning-based SC-ResNet50 (Skip Connection-Residual Network), which localizes the COVID-19 region in CXR images. In addition, a deep learning-based Robust Feature Neural Network (RFNN) is used to enhance the extraction of features from segmented CXR images. However, the early characteristics include joint COVID-19, pneumonia bacterial, viral, and normal properties. As a result, traditional approaches have failed to distinguish between different classes of disease-based features. The unique features of each class are extracted using the disease-specific feature separate attention mechanism (DSFSAM) in the RFNN. In addition, hybrid whale optimization algorithm (HWOA) based meta-heuristic is utilized to choose the best features in each class by using the hunting behavior of the whales. Finally, Q-learning-based Deep-Q-Neural Network (DQNN) is utilized to classify the various diseases, including COVID-19, pneumonia bacterial, viral, and normal, from CXR images. The procedural steps involved in the proposed MCSC-Net are given in Table 1.
Table 1
Procedural steps involved in the proposed MCSC-Net algorithm
| Proposed MCSC-Net algorithm |
| •Input: Training dataset, test CXR image |
| •Output: Preprocessed output, Segmented output, and Classified outcome, |
| •Performance metrics set-1: PSNR, SSIM, MSE, Entropy, MAE, PCC |
| •Performance metrics set-2: SACC, SSEN, SSPE, SRE, SF1, SPR |
| •Performance metrics set-3: CACC, CSEN, CSPE, CRE, CF1, CPR |
| Step 1: Apply the CXR input image to the HMBF preprocessing to remove the noises and enhances the COVID-19 region. |
| Step 2: Segment the lung region using the deep learning-based SC-ResNet50 model, highlighting the disease-affected regions. |
| Step 3: Extract the disease-specific features using RFNN with the DSFSAM model, which correlates the classes such as COVID-19, normal, Pneumonia bacterial and viral and forms the robust features. |
| Step 4: Apply HWOA to extract individual optimal features of the disease. |
| Step 5: Perform the multi-class classification using the DQNN model, which classifies the classes such as COVID-19, normal, bacterial, and viral pneumonia. |
| Step 6: Estimate the performance metrics set-1 such as PSNR, SSIM, MSE, and VQIM using preprocessed output. |
| Step 7: Estimate the performance metrics set-2 such as SACC, SSEN, SSPE, SRE, SF1, and SPR using segmented output. |
| Step 8: Estimate the performance metrics set-3 such as CACC, CSEN, CSPE, CRE, CF1, and CPR for 4, 3, and 2-class models. |
3.1Preprocessing
Noise removal is a significant process in CXR recognition to improve classification accuracy. MCSC-Net employs the Hybrid Median Bilateral Filter (HMBF) algorithm to remove noise in the given image. The input image dimension is 256×256. The proposed HMBF algorithm removes noise and sharpens the image effectually. In addition, it also maintains the fine details of the image. The proposed HMBF algorithm removes the universal noises from the given image, such as impulse and Gaussian. In the HMBF approach, the noisy pixel is identified using the Sorted Quadrant Median Vector (SQMV) algorithm. It preserves the important features of the CXR image, which are edges and texture information.
The functional blocks of the HMBF-based noise removal are depicted in Fig. 4. The four sequential blocks in HMBF are Adaptive Median Filter (AMF), Edge detector, Noise detector, and Switching Bilinear Filter (SBF). The operation of each sequential block present in the proposed HMBF is demonstrated in the following subsections.
Fig. 4
Proposed HMBF approach.
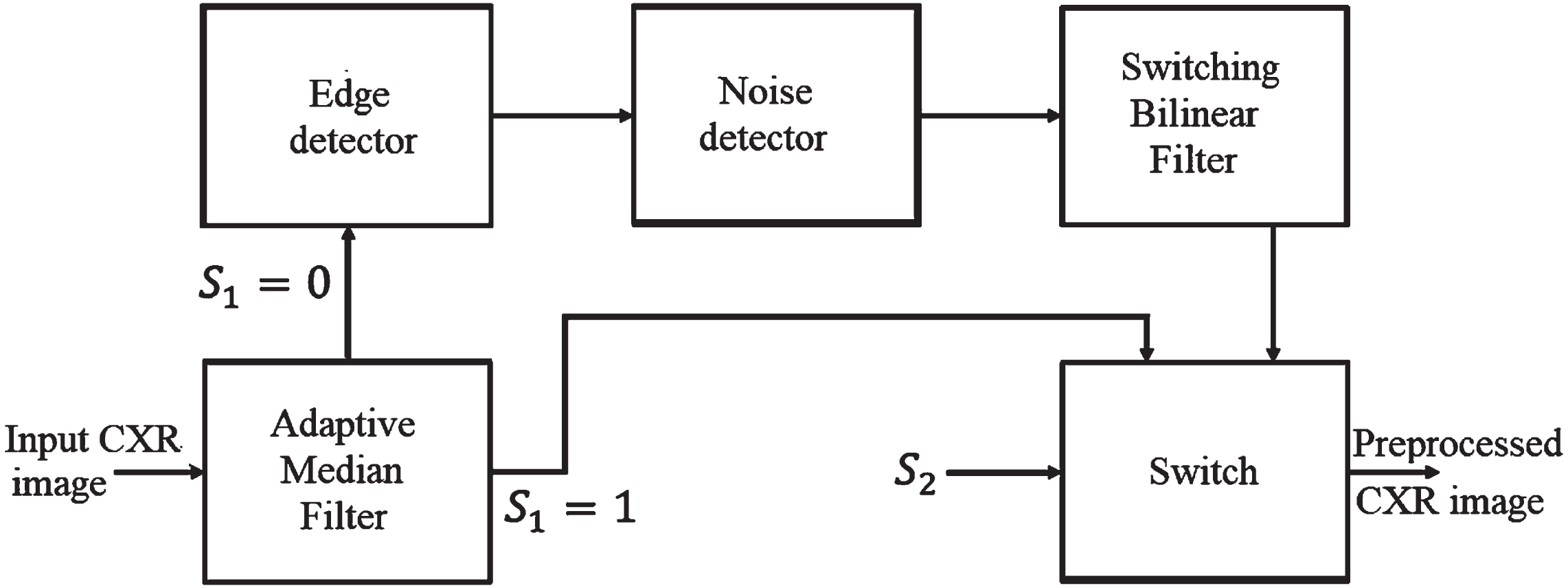
3.1.1Adaptive Median Filter (AMF)
Apply input noisy CXR image to the AMF filter, which is used to detect the contaminated pixels in the image. Most existing noise filtering algorithms employ constant window sizes such as 3*3 creates difficulties in differentiating the noisy and noise-free pixel that, results in blurriness in the output image. The proposed AMF algorithm adaptively modifies the window size to distinguish between noisy and noise-free pixels in the input image. Changing the image’s window size avoids blurriness and is more accessible to the process of noisy pixel detection. The AMF separates the noisy pixels and noise-less pixels based median switching condition of the switch by Equation (1).
(1)
3.1.2Edge detector
The edge detector is exploited to predict the edges of the current window accurately since it plays a vital role in CXR recognition. The edges are used to localize the noisy region.
3.1.3Noise detector
The noise detector is executed to select whether the current pixel is processed into the SBF Gaussian filter or SBF impulse filter. Assume that S1 and S2 are binary control signals, where AMF and noise detector creates S1 and S2. The image is filtered based on Equation (2).
(2)
3.1.4Switching Bilinear Filter with SQMV
The Switching Bilinear Filter (SBF) adaptively shifts its mode based on the results obtained from the noise detector, and the Sorted Quadrant Median Vector (SQMV) scheme is utilized to estimate the optimum median since the window size is adaptively changed. The SMQV finds the noisy pixel by finding the difference between the current pixel and the reference median pixel. If the difference between the current and reference median filter is large, then it is deliberated as the noisy pixel. Let assume fnd (i, j) indicates the current pixel and fnd (i + s, j + t) indicates the pixels in a (2N + 1) × (2N + 1) window adjacent to the fnd (i, j). The output (O (i, j)) from the SBF filter is obtained with Equation (3).
(3)
3.2Segmentation
The segmentation is the process of localizing the COVID-19 region from CXR images, and it also identifies the spatial coordinates of COVID-19. Here, the segmentation operation is accomplished to increase the image quality and reduce the CXR image’s adverse effects. The segmentation operation is performed by enhancing the COVID-19 region illumination, which is normalized using the SC-ResNet50-based transfer learning model. Figure 5 represents the proposed SC-ResNet50 model’s block diagram. With the help of the residual convolution module (RCM) and identity mapping module (IMM), the SC-ResNet50 effectively addresses the issue of fading gradients. Each layer of the IMM and RCM contributes to mapping the residual model, which is used in this module to connect the input and output.
Fig. 5
Proposed SC-ResNet50 (a) segmentation process, (b) IMM, (c) RCM.
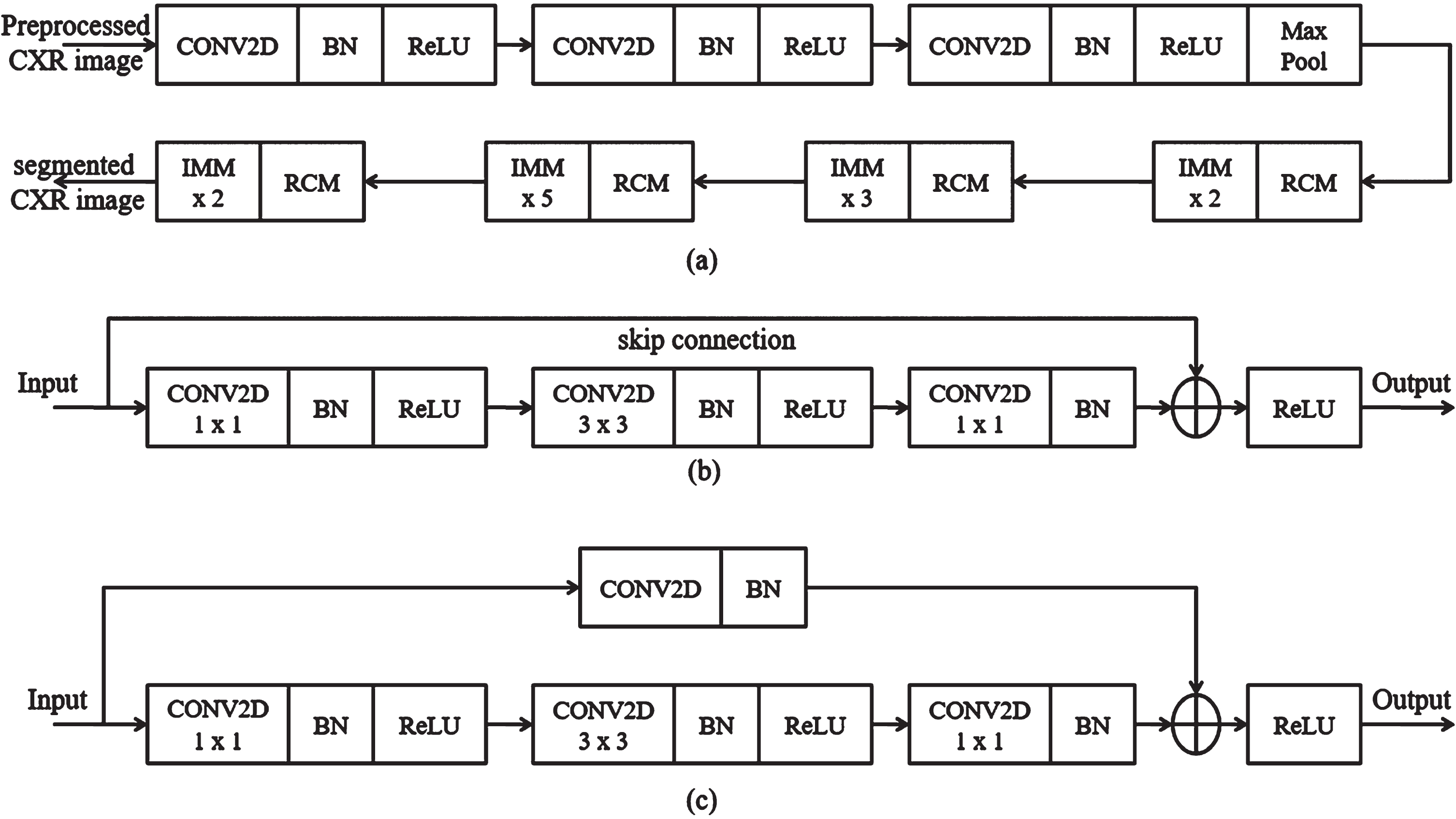
3.2.1Identity Mapping Module (IMM)
The proposed IMM layers improve the luminance and contrast of the image adaptively. The IMM layers establish dual gamma correction to improve the dark areas of the CXR image. In dual gamma correction, the first gamma correction is performed using convolution layers (CONV2D) to boost the image block’s entire luminance. Second, gamma correction is employed using a batch normalization (BN) layer to adjust the contrast of the dark regions in the image. It is performed to avoid the over-contrast enhancement result of the first gamma correction. The IMM model adaptively changes the clip points for the CXR image, which is set on the dynamic range of each block in the image to identify the COVID-19 region. The output (β) of the IMM is given in Equation (4).
(4)
In the above equation, in each block p indicates the number of the pixels, and dr means the dynamic range of the same block. τ and α weight regulating parameters for dr and entropies; further, σ is referred to as the standard deviation of the block, Av points out the mean, and c is the small value to avoid division by 0. Here, R is defined as the entire dynamic range of the image. Here, gmax signifies the maximum pixel value of the image.
3.2.2Residual convolution module
The dual gamma correction is achieved by introducing the Residual Convolution Module (RCM), which is performed after the completion of the clip point settings. The RCM model’s first gamma correction defines the weight (We) for global gray levels of the image blocks using Equation (5).
(5)
The Grmax designates the maximum gray value of the image, and Grref is referred to as the gray reference value of the image. The first (γ1) and second gamma (γ2) corrections are defined in Equations (6) and (7) respectively.
(6)
(7)
3.3Feature extraction
Multiple features are extracted from the segmented region using the Robust Feature Neural Network (RFNN) approach, and these particular disease-specific features are then extracted. The COVID-19, pneumonia bacterial, viral, and normal types play a vital role in classification operation because features of each disease type are dissimilar to others. Thus, the proposed deep learning-based RFNN model can extract the disease-specific and dependent features from segmented CXR images. The RFNN is capable of texture, shape, and spatial features. The RFNN descriptor is one of the feature descriptors in computer vision technology, which is partially inspired by the CNN descriptor. The deep learning models are popular for their computational speed and robustness to the illumination, scale, and rotation variations. Figure 6 shows the layer-wise structure of the RFNN model, which contains the nine series bocks for extracting the deep features. Further, RFNN encompasses four sequential layers in each block, such as feature extraction using CONV2D, DSFSAM, MaxPooling of features, and feature generation. Features can occasionally be single values that store the information in a pixel and are statical attributes in a vector format.
Fig. 6
RFNN feature extraction.
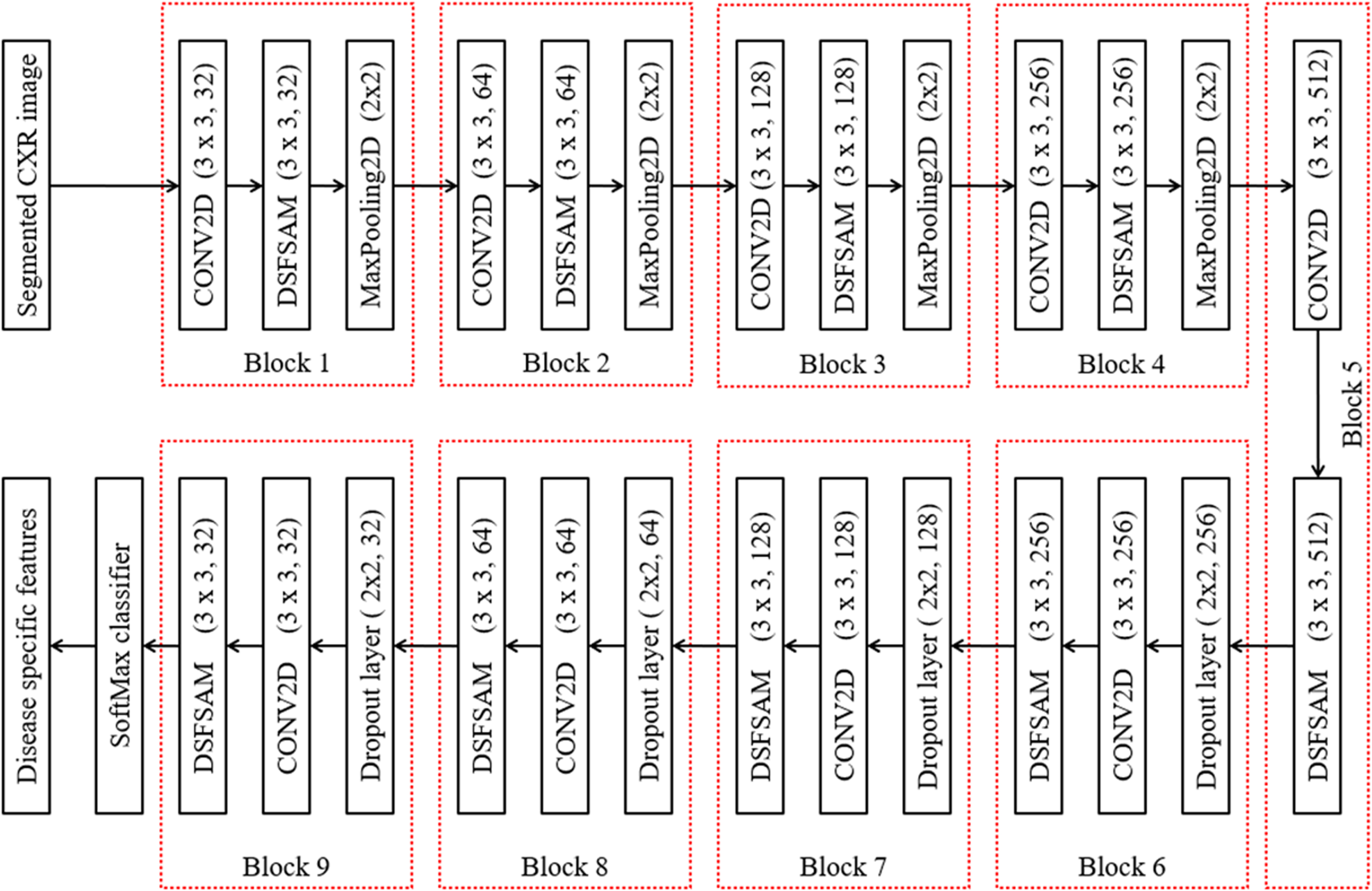
3.3.1Feature extraction using CONV2D
The RFNN utilizes square-shaped filters as an approximation of Gaussian smoothing, whereas CNN uses cascaded filters to estimate the scale-invariant features. The summation of all the pixels that exist in the segmented CXR image at location l = (m, n) in a rectangular region is known as integral images given by the Equation (8).
(8)
3.3.2Disease-specific feature separate attention mechanism
To find the disease-specific reference points, the RFNN algorithm uses the disease-specific feature separate attention mechanism (DSFSAM) detector based on the determinants of the Hessian matrix. The determinant of the Hessian matrix is used to measure the local variations around the point, and the points are selected where this determinant is high. The Hessian matrix is determined by using Equation (9).
(9)
(10)
The Hessian matrix determinant at different scales is indicated by Equation (11)
(11)
(12)
(13)
To determine the gradient of the disease-specific characteristics in either a horizontal or vertical direction, the DSFSAM filters are used. The following three points are used to assign the interest points in a dominating direction to produce candidate key points that are rotational invariant.
• The gradient of each disease-specific feature in the scale space is measured using the DSFSAM response.
• The window is rotated at a 60-degree angle around the circle’s center to produce the six vectors.
• The candidate for the critical point has the direction with the highest summation.
MaxPooling of features is used to eliminate the repeated disease-specific features and efficiently generate the output feature matrix.
3.3.3Feature generation
The square or box filter is rotated toward the key direction after selecting the neighborhood’s points for the feature generation. There are several smaller regions within the square region. For frequently spaced key points in each sub-region, DSFSAM is employed, and Equation (14) generates a four-dimensional vector.
(14)
3.4Feature selection using HWOA
The Whale Optimization technique developed by S. Mirjalili and A. Lewis [45], it is an efficient evolutionary optimization approach that may be used to find the best solution. Whales are highly intelligent creatures that prefer to live in groups rather than alone. This is the most interesting section of the whales’ bodies. The seeking properties of whales allow for the most optimum solutions to be found using the WOA approach. Walruses have a unique hunting technique in that they like to catch little fish that are near to the surface of the water. The hybrid whale optimization algorithm (HWOA) is constructed by combining the WOA with opposition-based learning (OBL) [25]. The proposed HWOA flowchart is shown in Fig. 7, which is a new and efficient optimization technique. The steps involved in the proposed HWOA are presented in Table 2. It helps to increase the convergence speed and the accuracy of solutions. The OBL approach selects an opposite number and places it at the candidate solution’s mirror location. Because the opposite number is extremely close to the random number that leads to a solution, algorithms utilize less space while converging the answer to the problem. The opposite population is more likely to come up with a global solution when compared to a random population. In the following mathematical explanations, the HWOA may be expressed in various phases such as encircling, hunting, targeting prey, and exploration.
Fig. 7
Proposed HWOA flowchart.
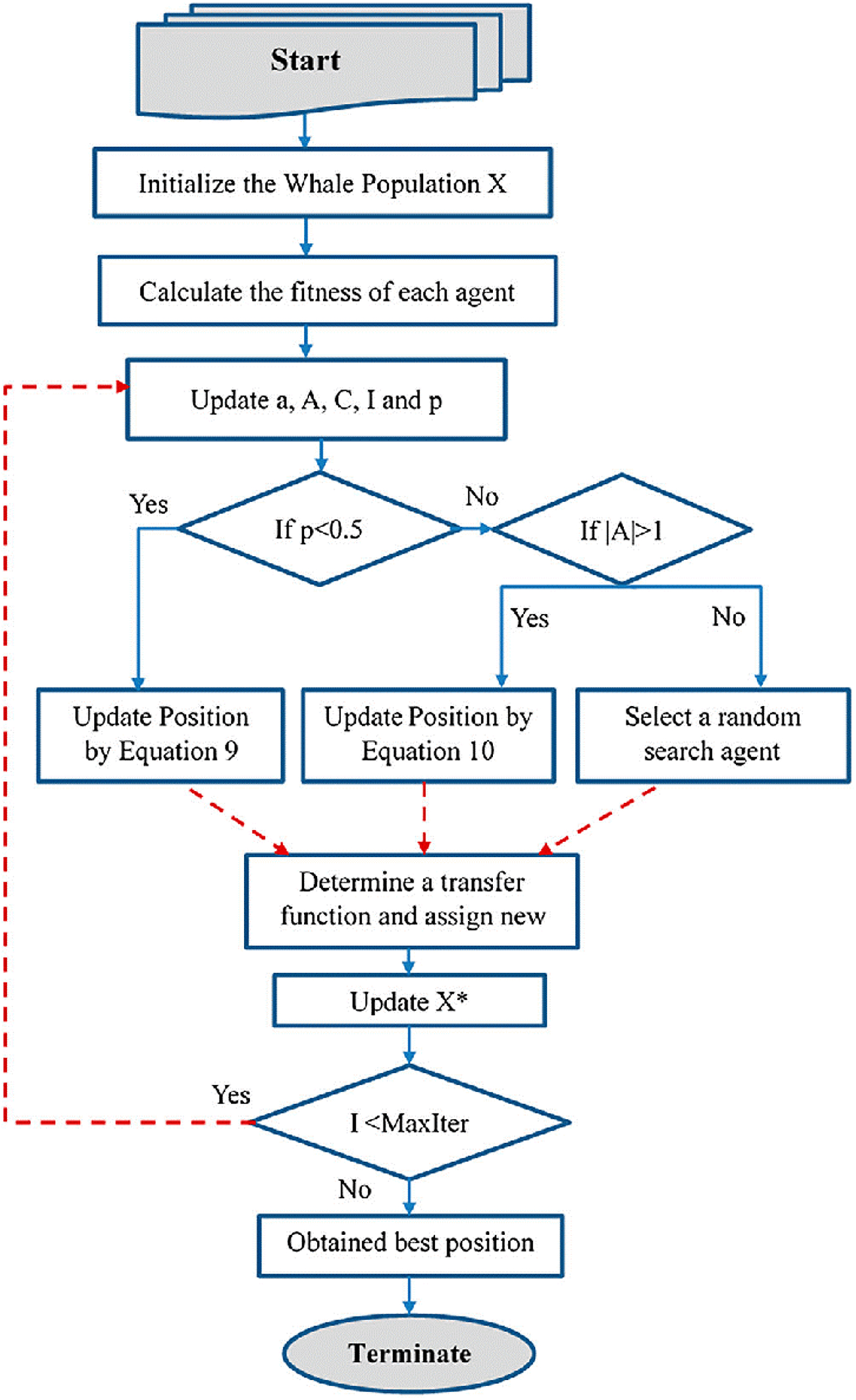
Table 2
Steps involved in the proposed HWOA
| Steps involved in the proposed HWOA |
| •Input: RFNN extracted features. |
| •Output: HWOA-based disease-specific features. |
| Step 1: Initialize the solutions according to population size. |
| Step 2: Create an opposite population; the independent variables of every solution are updated. |
| Step 3: The power generation of the Nth thermal unit (dependent variable) is calculated |
| Step 4: Find the population’s fitness value and the opposite population’s results. |
| Step 5: Choose Np numbers of fittest value from population and oppositional population sets. |
| Step 6: Fittest values are shorted in the form of finest to worst. |
| Step 7: Some solutions are kept as elite solutions |
| Step 8: Updates are being made to the independent variables of non-elite solutions. |
| Step 9: Again, calculate the power generation of the Nth thermal unit and fittest population set. |
| Step 10: Using jumping rate, the opposite population is generated from the new population |
| Step 11: Calculate opposition population fitness values. |
| Step 12: Np numbers of fittest values are taken from the current and the opposite population |
| Step 13: Repeat from Step8 for the next iteration |
3.4.1Encircling prey
The whales search for their prey by determining where it is most likely to be found and looking for its optimal position among the surrounding prey. All the other search agents closely track the position of the leading search agent, continually changing their own positions and searching around it. This potential option comes very close to being the best possible answer. The prey is encircled by encircling Equations (15–18) of the HWOA.
(15)
(16)
Where the whale position vector is
(17)
(18)
In this case,
3.4.2Bubble net hunting technique
A strategy for hunting humpback whales that uses bubble nets is based on the fact that these whales graze on the surface of the water. Whales attempt to communicate with one another via vocalization in order to create an efficient bubble net that allows them all to feed at the same time. Whales follow their prey by surrounding them with bubbles. They do this in a number of ways, including decreasing encircling and spiral position update. The condition for the mathematical formulation of the shrinking encircling agent is given by the Equation (19)
(19)
(20)
By creating and testing a mathematical model of humpback whales swimming around their prey, it is possible to characterize the mechanisms of the decreasing encircling and the spiral encircling methods. The Equation (21) gives two ways by which whales updated their position, with a 50 percent chance of selecting one of the two techniques being used in any given situation.
(21)
The shrinking and spiral encircling are hunting mechanisms in which a random number P is in [0, 1] range.
3.4.3Exploration phase (Search for prey)
To acquire the optimal solution, the prey, the search agent randomly adjusts its position in relation to the positions of other whales. The hunt for prey process may be expressed mathematically by the Equations (22, 23).
(22)
(23)
3.4.4Opposite number
The mirror positions of the proposed solution, which is the most crucial when considering the opposing variable into account. The opposing number (Xo) of candidate solution (X) with the interval [a, b] is a randomly generated in one-dimensional search space considering the following Equation (24).
(24)
The lowest and highest limits of the chosen search space are represented by α, and the symbols is b, respectively. Similar to the above, the Equation (25) may be used to describe the aforementioned statement in an n-dimensional search space:
(25)
The OBL is a novel approach to improving search ability and improving the solution accuracy of various optimization problems. To get the best solution, the HWOA searches for the solution in the opposite direction of specified values which is most likely to be nearer to a random number. The best features are formed by OBL, which is based on opposition-based generation jumping and opposition-based initialization.
3.5Classification
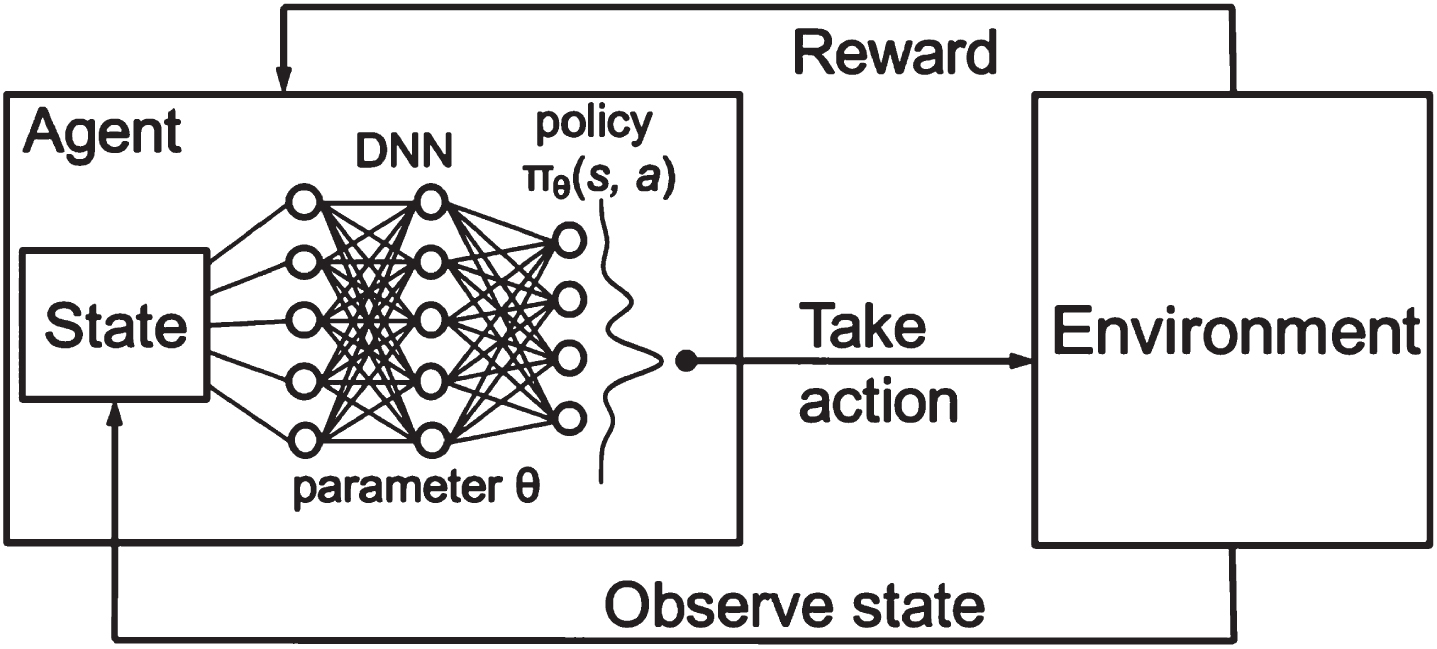
The HWOA-based optimal features are applied to the Deep-Q-Neural Network (DQNN) for classification. The proposed DQNN classifies texture features by implementing the Q-descriptor. The reason for selecting DQNN for texture feature extraction is that it performs better in extracting features from each image using different layers. The DQNN descriptor comprises three significant layers: DNN, Environment, and state layer, as depicted in Fig. 8. The Deep Neural Network (DNN) layer collects input from the input layer to derive the input image features. The convolution layer utilizes a number of filters to extract high-level features from the given image. The convolutional layer uses a collection of trainable filters to create the feature map. A total of six filters are used to create six feature maps from the input image. The feature map acquired from the individual filter is convolved through the whole image. Each feature map obtained from the filter signifies the precise features of the image. The DQNN operation is performed, which combines the two different functions to generate a third function. The DQNN operation is illustrated with Equation (26).
Fig. 8
DQNN descriptor.
(26)
where af designates the activation function, j signifies the specific convolution feature map, l exemplifies the layer in the CNN, fij represents the filter, bj is referred to as the feature map bias, and Ml is defined as the selection of feature map.
The environment layer is utilized to accomplish down sampling operation in the DQNN algorithm. The pooling operation is accomplished to diminish the spatial size representation and the volume of parameters and computations in the network. It functions on each feature map individually. The pooling environment operation is expressed with Equation (27).
(27)
where
4Results and discussions
This section discusses the results of the subjective and objective analyses in detail. The performance of the proposed method is compared with that of the currently utilized conventional approaches using three separate sets of performance metrics. All the methods considered the same dataset for evaluating the performance of the system. The segmentation and classification of proposed methods are estimated for 2-class, 3-class, and 4-class from CXR images. Here, 2-class models contain the COVID-19 and non-COVID-19 classification. Then, a 3-class model contains normal, COVID-19, and Pneumonia classification. Finally, the 4-class model contains normal, COVID-19, Pneumonia viral, and Pneumonia bacterial classification.
4.1Dataset
The dataset used in this work is collected from COVIDx CRX-2 [29], which is a publicly available kaggle repository. A total of 5371 posterior-to-anterior CXR scans from the thorax to the abdomen are taken from the repository. All of these CXR scans are a composite of an upgraded version of multiple previous datasets combined into a single final image. COVID-19 is a relatively new illness; as a result, the number of images belonging to this class is limited, which may lead to overfitting in prediction models. The collected dataset in this work contains 1500 images from the COVID-19 affected, 1500 of normal lungs and 1100 of pneumonia bacterial, and 1271 of pneumonia viral. In order to minimize overfitting, data augmentation was avoided. Data augmentation to produce more CXRs from 1500 existing CXR images of the COVID19 class could lead to a loss of generalization because of the large difference in data. As a result, class weights have been developed to address the class imbalance. The dataset is divided into 80% for training set and 20% for testing set. The dataset is split into two sets randomly by the Monte Carlo cross-validation. It was decided to use an intuitive strategy to determine the class weights to strongly penalize the loss that comes with the inaccurate categorization of the COVID-19 image. The detailed organization of the dataset used in this work for multiple segmentation and classification models is given in Table 3.
Table 3
Organization of dataset
| Model | COVID-19 CXRs | Normal CXRs | Pneumonia viral CXRs | Pneumonia bacterial CXRs |
| 4-Class | 1500 | 1500 | 1271 | 1100 |
| 3-Class | 1500 | 1500 | 2371 | |
| 2-Class | 1500 | 3871 |
4.2Subjective evaluation
The subjective evaluation gives the visual performance of the proposed method concerning preprocessing and segmentation methods used in this work. The preprocessing outputs of various methods such as Gabor filter, Wiener filter, histogram equalization, and proposed HMBF are shown in Fig. 9.
Fig. 9
Preprocessing performance evaluation (a) Input (b) Gabor filter (c) Wiener filter (d) Histogram Equalization (e) Proposed HMBF.
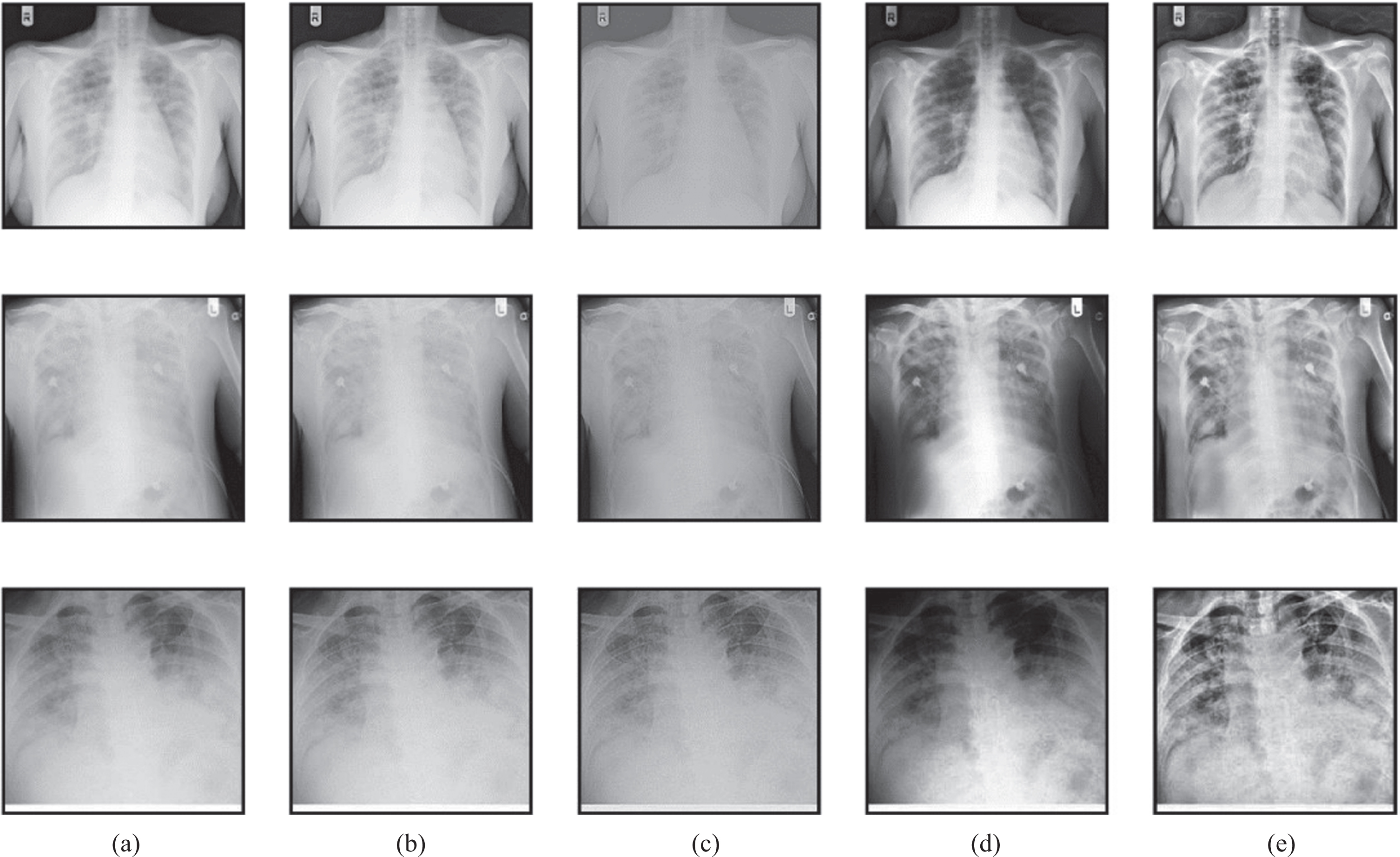
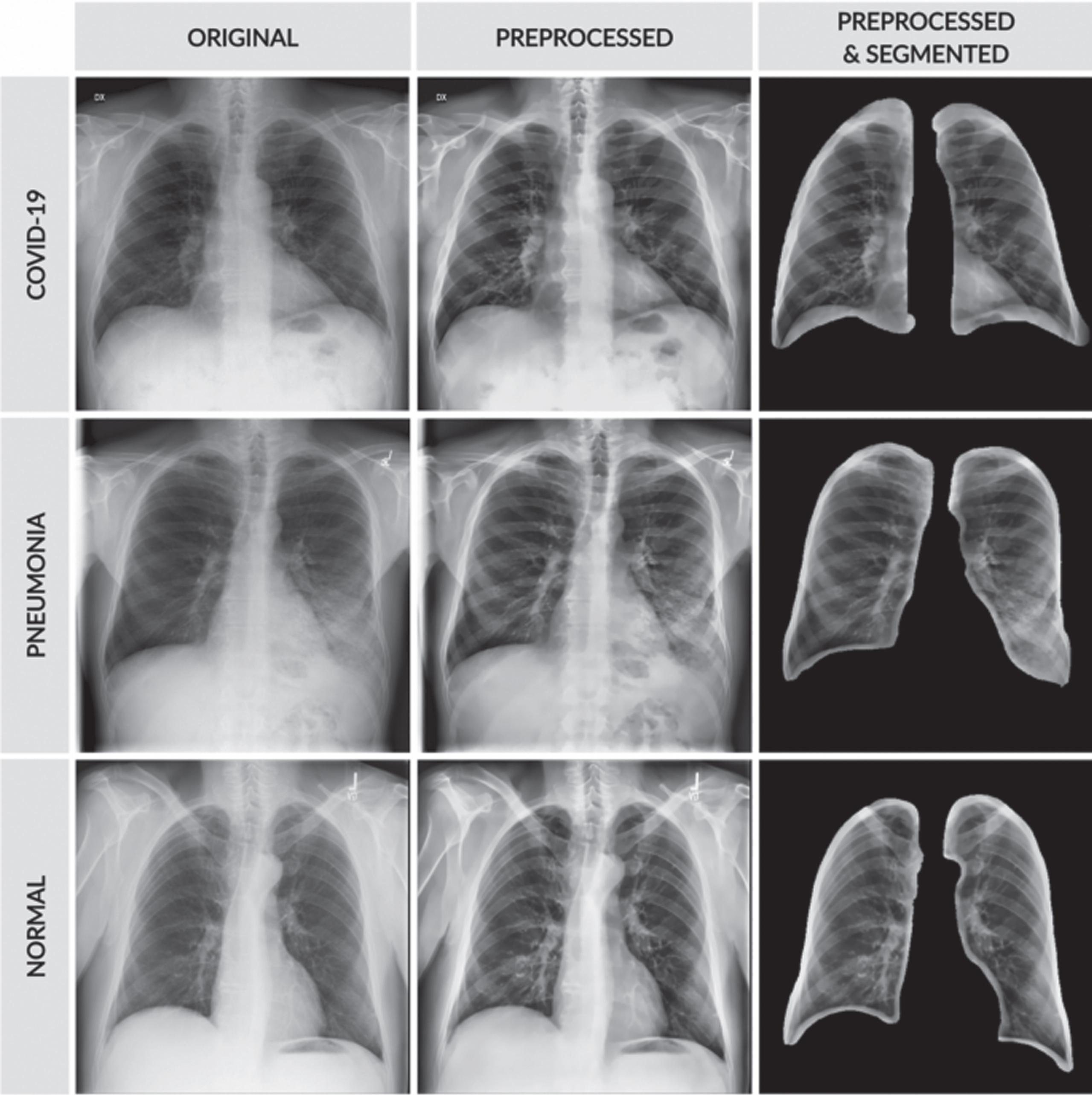
The input image in the first row is the COVID-19, the input image in the second row is pneumonia bacterial, and the input image in the third row is pneumonia viral. The proposed HMBF method perfectly enhances the input image with effective localization of disease-affected region visibility compared to the conventional approaches. The performance of SC-ResNet50 Segmentation with HMBF in preprocessing is shown in Fig. 10. Initially, the disease-affected CXR input images are applied to HMBF preprocessing, which enhances the disease-affected region. The SC-ResNet50 is developed to use the binary mask multiplied with the input image to generate the segmented region.
Fig. 10
SC-ResNet50 segmented outputs.
4.3Objective evaluation
Utilizing three separate sets of objective metrics, the objective evaluation compares the proposed method’s preprocessing, segmentation, and classification performance with that of existing conventional approaches.
4.3.1Preprocessing performance evaluation
The performance of the proposed preprocessing is compared with the existing preprocessing techniques with respect to Peak signal to noise ratio (PSNR), Structural similarity index metric (SSIM), Mean square error (MSE), Entropy, Pearson correlation coefficient (PCC), and Mean absolute error (MAE).
The performance comparison of the proposed HMBF preprocessing method with conventional approaches such as the Gabor filter, Wiener filter, and Histogram Equalization is given in Table 4. The proposed preprocessing method outperformed all performance criteria because it effectively visualizes the area impacted by the disease.
Table 4
Performance evaluation of proposed HMBF preprocessing method
| Method | PSNR (dB) | SSIM | MSE | Entropy | PCC | MAE |
| Gabor filter | 24.51 | 0.59 | 0.0825 | 34.11 | 0.5805 | 0.1462 |
| Wiener filter | 31.72 | 0.79 | 0.0601 | 43.75 | 0.7384 | 0.1083 |
| Histogram Equalization | 35.12 | 0.84 | 0.0244 | 48.14 | 0.8777 | 0.0990 |
| Proposed HMBF | 42.75 | 9.99 | 0.0180 | 53.02 | 0.9934 | 0.0451 |
4.3.2Segmentation performance evaluation
The performance of the proposed SC-ResNet50 segmentation for CXR image segmentation is given in Table 5. The performance is provided with respect to the metrics such as segmentation accuracy (SACC), segmentation sensitivity (SSEN), segmentation specificity (SSPE), segmentation recall (SRE), segmentation F1-score (SF1), and segmentation precision (SPR). According to Table 5, the 4-class model outperformed the 3 and 2-class models in terms of segmentation performance.
Table 5
Performance evaluation of proposed SC-ResNet50 segmentation model
| Models | SACC | SSEN | SSPE | SRE | SF1 | SPR |
| 2-Class | 98.11 | 98.40 | 99.86 | 99.00 | 97.48 | 98.53 |
| 3-Class | 98.58 | 98.50 | 98.71 | 98.31 | 99.59 | 99.46 |
| 4-Class | 99.26 | 99.49 | 99.03 | 98.07 | 98.13 | 99.93 |
| Average | 98.65 | 98.79 | 99.20 | 98.46 | 98.4 | 99.30 |
The proposed 2-class SC-ResNet50 segmentation performance is compared with the conventional approaches such as DEFU-Net [28], FractalCovNet [23], and SegNet [10], as given in Table 6. The traditional models failed to localize the disease-affected region, so the performance of conventional models is lower than the proposed segmentation model.
Table 6
Performance comparison of 2-class segmentation models
| Models | SACC | SSEN | SSPE | SRE | SF1 | SPR |
| DEFU-Net [28] | 94.34 | 94.02 | 96.13 | 94.46 | 95.99 | 94.50 |
| FractalCovNet [23] | 96.65 | 96.22 | 97.70 | 96.32 | 96.22 | 96.87 |
| SegNet [10] | 93.85 | 98.02 | 88.84 | 98.02 | 94.56 | 91.33 |
| 2-class SC-ResNet50 | 98.11 | 98.40 | 99.86 | 99.00 | 97.48 | 98.53 |
The performance of the proposed 3-class SC-ResNet50 segmentation model is given in Table 7 and is compared with the existing conventional approaches such as DeepSDM [50], COVID-SDNet [40], and RANDGAN [41]. The proposed method accurately identified the lung region compared to existing models.
Table 7
Performance comparison of 3-class segmentation models
| Models | SACC | SSEN | SSPE | SRE | SF1 | SPR |
| DeepSDM [50] | 93.89 | 90.06 | 95.69 | 91.41 | 90.41 | 93.44 |
| SDNet [40] | 94.32 | 93.87 | 94.35 | 93.83 | 93.83 | 95.97 |
| RANDGAN [41] | 96.76 | 94.71 | 90.00 | 94.71 | 94.71 | 96.32 |
| 3-class SC-ResNet50 | 98.58 | 98.50 | 98.71 | 98.31 | 99.59 | 99.46 |
The performance of the proposed 4-class SC-ResNet50 segmentation model is given in Table 8 and is compared with the conventional approaches such as VAE-TTAM [21], SADNN [32], and MTDL [33]. The segmented outcome of the proposed approach accurately matched the ground truth, which caused improvement in the proposed method.
Table 8
Performance comparison of 4-class segmentation models
| Models | SACC | SSEN | SSPE | SRE | SF1 | SPR |
| VAE-TTAM [21] | 93.41 | 93.51 | 95.45 | 93.59 | 93.22 | 94.31 |
| SADNN [32] | 93.61 | 96.13 | 95.63 | 96.11 | 94.38 | 95.68 |
| MTDL [33] | 97.85 | 97.19 | 97.20 | 97.06 | 96.43 | 96.71 |
| 4-class SC-ResNet50 | 99.26 | 99.49 | 99.03 | 98.07 | 98.13 | 99.93 |
The performance of the individual segmentation models is compared and listed in Tables 6 to 8. But there are few models which are developed to perform multi-class segmentation. Therefore, the performance of the proposed SC-ResNet50 segmentation model is compared with the conventional existing multi-class segmentation models such as U-Net [22], TL-CNN [7], and DenseCapsNet [24] in Table 9. The effectiveness of the suggested segmentation method is examined here with respect to the individual 4-class, 3-class, and 2-class models of the existing methodologies. As a result of its successful localization of the disease-affected boundary, the suggested SC-ResNet50 segmentation model of this work outperformed the multi-class segmentation models presented in Table 9 in terms of performance.
Table 9
Performance comparison of multi-class segmentation models
| Class | Models | SACC | SSEN | SSPE | SRE | SF1 | SPR |
| 4-Class | U-Net [22] | 94.32 | 94.34 | 94.81 | 93.33 | 93.48 | 92.13 |
| TL-CNN [7] | 96.85 | 95.24 | 96.40 | 95.82 | 94.34 | 94.64 | |
| DenseCapsNet [24] | 97.79 | 97.95 | 97.47 | 97.97 | 96.80 | 95.98 | |
| SC-ResNet50 | 99.26 | 99.49 | 99.03 | 98.07 | 98.13 | 99.93 | |
| 3-Class | U-Net [22] | 93.12 | 93.53 | 94.37 | 93.30 | 93.75 | 94.02 |
| TL-CNN [7] | 95.43 | 94.30 | 95.35 | 95.67 | 94.65 | 95.23 | |
| DenseCapsNet [24] | 97.51 | 95.98 | 96.57 | 96.77 | 96.14 | 96.09 | |
| SC-ResNet50 | 98.58 | 98.50 | 98.71 | 98.31 | 99.59 | 99.46 | |
| 2-Class | U-Net [22] | 93.36 | 94.41 | 95.29 | 93.79 | 93.08 | 95.37 |
| TL-CNN [7] | 95.89 | 95.91 | 96.00 | 96.16 | 95.77 | 96.41 | |
| DenseCapsNet [24] | 97.64 | 97.96 | 97.37 | 97.94 | 97.77 | 97.66 | |
| SC-ResNet50 | 98.11 | 98.40 | 99.86 | 99.00 | 97.48 | 98.53 |
4.3.3Classification performance evaluation
The performance of various classification models is compared with the proposed MCSC-Net with respect to the metrics such as classification accuracy (CACC), classification sensitivity (CSEN), classification specificity (CSPE), classification recall (CRE), classification F1-Score (CF1), and classification precision (CPR). The proposed MCSC-Net performance with respect to each class, such as 4-class, 3-class, and 2-class classification, is compared in Table 10. The performance evaluation metrics listed in Table 9 show that the proposed MCSC-Net shows superiority with an average classification of 99.16% over all classes. Also, it shows that the proposed MCSC-Net classification resulted in more classification accuracy in 4-class classification compared to 3-class and 2-class classification.
Table 10
Performance evaluation of proposed MCSC-Net classification models
| Models | CACC | CSEN | CSPE | CRE | CF1 | CPR |
| 2-Class | 99.0911 | 99.384 | 99.8586 | 99.9900 | 98.4548 | 99.5153 |
| 3-Class | 99.1658 | 99.485 | 99.6971 | 99.2931 | 99.5859 | 99.4546 |
| 4-Class | 99.2526 | 99.487 | 99.0203 | 99.0507 | 99.1113 | 99.9293 |
| Average | 99.1698 | 99.452 | 99.5253 | 99.4446 | 99.0506 | 99.6330 |
The performance of the proposed 2-class MCSC-Net classification models with state-of-the-art other existing 2-class classification conventional models such as ASSOA-ResNet [17], QRSMA-CNN [42], and mWOAPR-SVM [43] are compared in Table 11. The conventional models failed to extract the detailed features, and due to obscene segmentation methods, they achieved low classification accuracy than the proposed MCSC-Net model.
Table 11
Performance comparison of 2-class classification models
| Models | CACC | CSEN | CSPE | CRE | CF1 | CPR |
| ASSOA-ResNet [17] | 92.169 | 92.4561 | 92.9115 | 92.4066 | 94.9509 | 92.9016 |
| QRSMA-CNN [42] | 93.3966 | 93.0798 | 95.1687 | 93.5154 | 95.0301 | 93.555 |
| mWOAPR-SVM [43] | 95.6835 | 95.2578 | 96.723 | 95.3568 | 95.2578 | 95.9013 |
| 2-class MCSC-Net | 99.0911 | 99.384 | 99.8586 | 99.9900 | 98.4548 | 99.5153 |
The performance of the proposed 3-class MCSC-Net classification model is compared with state-of-the-art other existing conventional 3-Class classification models such as InceptionV3 [38], FOMPA-CNN [8], and MobileNetV3 [34] in Table 12. These conventional methods failed to select the disease-specific features and did not utilize the preprocessing operations, which caused the reduction of overall classification performance as compared to the proposed MCSC-Net model.
Table 12
Performance comparison of 3-class classification models
| Models | CACC | CSEN | CSPE | CRE | CF1 | CPR |
| InceptionV3 [38] | 92.169 | 92.4561 | 92.9115 | 92.4066 | 94.9509 | 92.9016 |
| FOMPA-CNN [8] | 93.3966 | 93.0798 | 95.1687 | 93.5154 | 95.0301 | 93.555 |
| MobileNetV3 [34] | 95.6835 | 95.2578 | 96.723 | 95.3568 | 95.2578 | 95.9013 |
| 3-class MCSC-Net | 99.1658 | 99.485 | 99.6971 | 99.2931 | 99.5859 | 99.4546 |
The performance of the proposed 4-class MCSC-Net classification model is compared in Table 13 with existing 4-class classification models such as OptCoNet [46], PSO-VGG19 [15], and GWOA-CNN [19]. The proposed MCSC-Net was implemented with preprocessing, segmentation, feature extraction, and classification stages, which causes improved classification performance in convention with existing approaches.
Table 13
Performance comparison of 4-class classification models
| Models | CACC | CSEN | CSPE | CRE | CF1 | CPR |
| OptCoNet [46] | 92.4759 | 92.5749 | 94.4955 | 92.6541 | 92.2878 | 93.3669 |
| PSO-VGG19 [15] | 93.6739 | 95.1687 | 94.6737 | 95.1489 | 93.4362 | 94.7232 |
| GWOA-CNN [19] | 96.8715 | 96.2181 | 96.228 | 96.0894 | 95.4657 | 95.7429 |
| 4-class MCSC-Net | 99.2526 | 99.484 | 99.0203 | 99.0507 | 99.1113 | 99.9293 |
The comparisons provided in Tables 11 to 13 of the proposed MCSC-Net classification are based on the individual class. There are few conventional existing multi-class classification models on CXR images. The performance of the proposed multi-class MCSC-Net is compared with the existing multi-class state of art classification models such as IG-CNN [9], WOA-GAN [47], and BAT-MLP [44] in Table 14. The proposed multi-class MCSC-Net is capable of extracting the disease-dependent features and also selecting the disease-specific features, which causes improvement in performance for all classes.
Table 14
Performance comparison of multi-class MCSC-Net classification models
| Class | Models | CACC | CSEN | CSPE | CRE | CF1 | CPR |
| 4-Class | IG-CNN [9] | 93.376 | 93.3966 | 93.8619 | 92.3967 | 92.5452 | 91.2087 |
| WOA-GAN [47] | 95.8815 | 94.2876 | 95.436 | 94.8618 | 93.3966 | 93.6936 | |
| BAT-MLP [44] | 96.8121 | 96.9705 | 96.4953 | 96.9903 | 95.832 | 95.0202 | |
| MCSC-Net | 99.2526 | 99.484 | 99.0203 | 99.0507 | 99.1113 | 99.9293 | |
| 3-Class | IG-CNN [9] | 92.1888 | 92.5947 | 93.4263 | 92.367 | 92.8125 | 93.0798 |
| WOA-GAN [47] | 94.4757 | 93.357 | 94.3965 | 94.7133 | 93.7035 | 94.2777 | |
| BAT-MLP [44] | 96.5349 | 95.0202 | 95.6043 | 95.8023 | 95.1786 | 95.1291 | |
| MCSC-Net | 99.1658 | 99.485 | 99.6971 | 99.2931 | 99.5859 | 99.4546 | |
| 2-Class | IG-CNN [9] | 92.4264 | 93.4659 | 94.3371 | 92.8521 | 92.1492 | 94.4163 |
| WOA-GAN [47] | 94.9311 | 94.9509 | 95.04 | 95.1984 | 94.8123 | 95.4459 | |
| BAT-MLP [44] | 96.6636 | 96.9804 | 96.3963 | 96.9606 | 96.7923 | 96.6834 | |
| MCSC-Net | 99.0911 | 99.384 | 99.8586 | 99.9900 | 98.4548 | 99.5153 |
5Conclusion
The proposed MCSC-Net model is developed by combining the Deep learning, Transfer learning, Bio-optimization algorithm, and Q-learning models, which resulted in a hybrid approach and held the advantages of all individual models in CXR image classification. To enhance the CXR images and disease-affected regions in CXR images by noise removal, a preprocessing HMBF was developed. Then, the preprocessed CXR images were segmented by the transfer learning-based SC-ResNet50 model, which effectively localizes the lung-based disease-affected region. Further, a deep learning-based RFNN model is developed to extract the disease-specific features related to the normal, COVID-19, Bacterial and Viral Pneumonia classes separately. The developed RFNN unutilized the DSFSAM model for non-overlapping separation of class-specific features. The optimal and best features are selected from RFNN using HWOA based Bio-optimization method. Finally, Q-learning-based DQNN was used to perform the multi-class classification operations. In this SC-ResNet50-based segmentation and DQNN-based classification, operations are carried out for 2-claas, 3-class, and 4-class classifications separately. The implemented result demonstrates that, when compared to the traditional current multi-class models in CXR image classification, the proposed MCSC-Net model with the proposed preprocessing, segmentation and classification resulted in improved performance. Additionally, the COVID-19 in CXR images is effectively classified by the MCSC-Net. This study may be expanded to incorporate other optimization techniques for reducing system losses.
Funding
None.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
No studies involving humans or animals have been reported by any of the authors in this article.
References
[1] | Ibrahim A.U. , et al., Pneumonia classification using deep learning from chest X-ray images during COVID-19, Cognitive Computation (2021), 1–13. doi: 10.1007/s12559-020-09787-5 |
[2] | Shelke A. , et al., Chest X-ray classification using deep learning for automated COVID-19 screening, SN Computer Science 2: (4) ((2021) ), 1–9. doi: 10.1007/s42979-021-00695-5 |
[3] | Ismael A.M. and Sengür A. , Deep learning approaches for COVID-19 detection based on chest X-ray images, Expert Systems with Applications 164: ((2021) ), 114054. doi: 10.1016/j.eswa.2020.114054 |
[4] | Majid A. , et al., COVID19 classification using CT images via ensembles of deep learning models, Computers, Materials and Continua 69: (1) ((2021) ), 319–337. doi: 10.32604/cmc.2021.016816 |
[5] | Amyar A. , et al., Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: Classification and segmentation, Computers in Biology and Medicine 126: ((2020) ), 104037. doi: 10.1016/j.compbiomed.2020.104037 |
[6] | Waheed A. , et al., CovidGAN: Data augmentation using auxiliary classifier GAN for improved covid-19 detection, IEEE Access 8: ((2020) ), 91916–91923. doi: 10.1109/ACCESS.2020.2994762 |
[7] | Narin A. , Kaya C. and Pamuk Z. , Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks, Pattern Analysis and Applications 24: (3) ((2021) ), 1207–1220. doi: 10.1007/s10044-021-00984-y |
[8] | Sahlol A.T. , et al., COVID-19 image classification using deep features and fractional-order marine predators algorithm, Scientific Reports 10: ((2020) ), 15364. doi: 10.1038/s41598-020-71294-2 |
[9] | Al-zubidi A. , et al., Mobile application to detect covid-19 pandemic by using classification techniques: Proposed system, International Journal of Interactive Mobile Technologies 15: (16) ((2021) ), 34–51. doi: 10.3991/ijim.v15i16.24195 |
[10] | Gopatoti A. and Vijayalakshmi P. , Optimized chest X-ray image semantic segmentation networks for COVID-19 early detection, Journal of X-Ray Science and Technology 30: (3) ((2022) ), 491–512. doi: 10.3233/XST-211113 |
[11] | Gopatoti A. and Vijayalakshmi P. , CXGNet: A tri-phase chest X-ray image classification for COVID-19 diagnosis using deep CNN with enhanced grey-wolf optimizer, Biomedical Signal Processing and Control 77: ((2022) ), 103860. doi: 10.1016/j.bspc.2022.103860 |
[12] | Pirouz B. , et al., Investigating a serious challenge in the sustainable development process: Analysis of confirmed cases of COVID-19 (new type of coronavirus) through a binary classification using artificial intelligence and regression analysis, Sustainability 12: (6) ((2020) ), 2427. doi: 10.3390/su12062427 |
[13] | Stifanić D. , et al., Semantic segmentation of chest X-rayimages based on the severity of COVID-19 infected patients, EAI Endorsed Transactions on Bioengineering and Bioinformatics 1: (3) ((2021) ), e3. |
[14] | Ibrahim D.M. , Elshennawy N.M. and Sarhan A.M. , Deep-chest: Multi-classification deep learning model for diagnosing COVID-19, pneumonia, and lung cancer chest diseases, Computers in Biology and Medicine 132: ((2021) ), 104348. doi: 10.1016/j.compbiomed.2021.104348 |
[15] | Dias Júnior D.A. , et al., Automatic method for classifyingCOVID-19 patients based on chest X-ray images, using deep featuresand PSO-optimized XGBoost, Expert Systems with Applications 183: ((2021) ), 115452. doi: 10.1016/j.eswa.2021.115452 |
[16] | Hussain E. , et al., CoroDet: A deep learning based classification for COVID-19 detection using chest X-ray images, Chaos, Solitons & Fractals 142: ((2021) ), 110495. doi: 10.1016/j.chaos.2020.110495 |
[17] | El-Kenawy E.-S.M. , et al., Advanced meta-heuristics, convolutional neural networks, and feature selectors for efficient COVID-19 X-ray chest image classification, IEEE Access 9: ((2021) ), 36019–36037. doi: 10.1109/ACCESS.2021.3061058 |
[18] | Carvalho E.D. , et al., An approach to the classification of COVID-19 based on CT scans using convolutional features and genetic algorithms, Computers in Biology and Medicine 136: ((2021) ), 104744. doi: 10.1016/j.compbiomed.2021.104744 |
[19] | El-Kenawy E.-S.M. , et al., Novel feature selection and voting classifier algorithms for COVID-19 classification in CT images, IEEE Access 8: ((2020) ), 179317–179335. doi: 10.1109/ACCESS.2020.3028012 |
[20] | Shi F. , et al., Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID-19, IEEE Reviews in Biomedical Engineering 14: ((2020) ), 4–15. doi: 10.1109/RBME.2020.2987975 |
[21] | Cao F. and Zhao H. , Automatic lung segmentation algorithm on chest x-ray images based on fusion variational auto-encoder and three-terminal attention mechanism, Symmetry 13: (5) ((2021) ), 814. doi: 10.3390/sym13050814 |
[22] | Gaál G. , Maga B. and Lukács A. , Attention U-Net based adversarial architectures for chest x-ray lung segmentation, arXiv: 2003.10304 (2020). doi: 10.48550/arXiv.2003.10304 |
[23] | Munusamy H. , et al., FractalCovNet architecture for COVID-19 chest X-ray image classification and CT-scan image segmentation, Biocybernetics and Biomedical Engineering 41: (3) ((2021) ), 1025–1038. doi: 10.1016/j.bbe.2021.06.011 |
[24] | Quan H. , et al., DenseCapsNet: Detection of COVID-19 from X-ray images using a capsule neural network, Computers in Biology and Medicine 133: ((2021) ), 104399. doi: 10.1016/j.compbiomed.2021.104399 |
[25] | Tizhoosh H.R. , Opposition-based learning: A new scheme for machine intelligence, In 567 International Conference on Computational Intelligence for Modelling, Control and Automation and International Conference on Intelligent Agents, Web Technologies and Internet Commerce (CIMCA-IAWTIC’06 IEEE.), 2005, pp. 695–701. doi: 10.1109/CIMCA.2005.1631345 |
[26] | Shankar K. and Perumal E. , A novel hand-crafted with deep learning features based fusion model for COVID-19 diagnosis and classification using chest X-ray images, Complex & Intelligent Systems 7: (3) ((2021) ), 1277–1293. doi: 10.1007/s40747-020-00216-6 |
[27] | Teixeira L.O. , et al., Impact of lung segmentation on the diagnosis and explanation of COVID-19 in chest X-ray images, Sensors 21: (21) ((2021) ), 7116. doi: 10.3390/s21217116 |
[28] | Zhang L. , et al., Dual Encoder Fusion U-Net (DEFU-Net) for Cross-manufacturer Chest X-ray Segmentation, In 2020 25th International Conference on Pattern Recognition (ICPR) IEEE, 2021, pp. 9333–9339. doi: 10.1109/ICPR48806.2021.9412718 |
[29] | Wang L. , et al., COVID-Net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images, Scientific Reports 10: ((2020) ), 19549. doi: 10.1038/s41598-020-76550-z |
[30] | Monshi M.M.A. , Poon J. , et al., CovidXrayNet: Optimizing data augmentation and CNN hyperparameters for improved COVID-19 detection from CXR, Computers in Biology and Medicine 133: ((2021) ), 104375. doi: 10.1016/j.compbiomed.2021.104375 |
[31] | Horry M.J. , et al., COVID-19 detection through transfer learning using multimodal imaging data, IEEE Access 8: ((2020) ), 149808–149824. doi: 10.1109/ACCESS.2020.3016780 |
[32] | Kim M. and Lee B.-D. , Automatic lung segmentation on chest X-rays using self-attention deep neural network, Sensors 21: (2) ((2021) ), 369. doi: 10.3390/s21020369 |
[33] | Alom M.Z. , et al., COVID MTNet: COVID-19 detection with multi-task deep learning approaches, arXiv: 2004.03747 (2020). doi: 10.48550/arXiv.2004.03747 |
[34] | A. Elaziz M. , et al., Boosting COVID-19 image classification using MobileNetV3 and Aquila optimizer algorithm, Entropy 23: (11) ((2021) ), 1383. doi: 10.3390/e23111383 |
[35] | Vidal P.L. , et al., Multi-stage transfer learning for lung segmentation using portable X-ray devices for patients withCOVID-19, Expert Systems with Applications 173: ((2021) ), 114677. doi: 10.1016/j.eswa.2021.114677 |
[36] | Bharat Siva Varma P. , et al., SLDCNet: Skin lesion detection andclassification using full resolution convolutional network-baseddeep learning CNN with transfer learning, Expert Systems 39: (9) ((2022) ), e12944. doi: 10.1111/exsy.12944 |
[37] | Suppakitjanusant P. , Sungkanuparph S. , et al., Identifying individuals with recent COVID-19 through voice classification using deep learning, Scientific Reports 11: (1) ((2021) ), 1–7. doi: 10.1038/s41598-021-98742-x |
[38] | Bhowal P. , Sen S. and Sarkar R. , A two-tier feature selection method using Coalition game and Nystrom sampling for screening COVID-19 from chest X-Ray images, Journal of Ambient Intelligence and Humanized Computing (2021), 1–16. doi: 10.1007/s12652-021-03491-4 |
[39] | Serte S. and Demirel H. , Deep learning for diagnosis of COVID-19 using 3D CT scans, Computers in Biology and Medicine 132: ((2021) ), 104306. doi: 10.1016/j.compbiomed.2021.104306 |
[40] | Tabik S. , et al., COVIDGR dataset and COVID-SDNet methodology for predicting COVID-19 based on chest X-ray images, IEEE Journal of Biomedical and Health Informatics 24: (12) ((2020) ), 3595–3605. doi: 10.1109/JBHI.2020.3037127 |
[41] | Motamed S. , Rogalla P. and Khalvati F. , RANDGAN: Randomized Generative Adversarial Network for detection of COVID-19 in chest X-ray, Scientific Reports 11: (1) ((2021) ), 1–10. doi: 10.1038/s41598-021-87994-2 |
[42] | Nama S. , A novel improved SMA with quasi reflection operator: Performance analysis, application to the image segmentation problem of Covid-19 chest X-ray images, Applied Soft Computing 118: ((2022) ), 108483. doi: 10.1016/j.asoc.2022.108483 |
[43] | Chakraborty S. , et al., COVID-19 X-ray image segmentation by modified whale optimization algorithm with population reduction, Computers in Biology and Medicine 139: ((2021) ), 104984. doi: 10.1016/j.compbiomed.2021.104984 |
[44] | Pathan S. , Siddalingaswamy P.C. and Ali T. , Automated detection of covid-19 from chest x-ray scans using an optimized CNN architecture, Applied Soft Computing 104: ((2021) ), 107238. doi: 10.1016/j.asoc.2021.107238 |
[45] | Mirjalili S. and Lewis A. , The whale optimization algorithm, Advances in Engineering Software 95: ((2016) ), 51–67. doi: 10.1016/j.advengsoft.2016.01.008 |
[46] | Goel T. , et al., OptCoNet: An optimized convolutional neural network for an automatic diagnosis of COVID-19, Applied Intelligence 51: (3) ((2021) ), 1351–1366. doi: 10.1007/s10489-020-01904-z |
[47] | Goel T. , et al., Automatic screening of covid-19 using an optimized generative adversarial network, Cognitive Computation (2021). doi: 10.1007/s12559-020-09785-7 |
[48] | Ravi V. , et al., Deep learning-based meta-classifier approach for COVID-19 classification using CT scan and chest X-ray images, Multimedia Systems 28: ((2021) ), 1401–1415. doi: 10.1007/s00530-021-00826-1 |
[49] | WHO coronavirus (COVID-19) dashboard [Online]. Available: https://covid19.who.int/ |
[50] | Wang Y. , et al., DeepSDM: Boundary-aware pneumothorax segmentation in chest X-ray images, Neurocomputing 454: ((2021) ), 201–211. doi: 10.1016/j.neucom.2021.05.029 |
[51] | Karhan Z. and Akal F. , Covid-19 classification using deep learning in chest X-ray images, Medical Technologies Congress (TIPTEKNO) (2020), 1–4. doi: 10.1109/TIPTEKNO50054.2020.9299315 |




