Grape skin extract prevents UV irradiation induced DNA damage of normal human epidermal keratinocytes cells
Abstract
BACKGROUND:
There is concern that DNA damage may occur in skin cells due to UV irradiation. In this study, we investigated whether extracts from grape skin, which should be discarded, can suppress DNA damage caused by UV radiation.
OBJECTIVE:
This study aims to investigate the effectivity of GSE in diminishing UV-induced cytotoxicity in normal human epidermal keratinocytes (NHEK) cells.
METHODS:
The polyphenol content in GSE was carried out using the HPLC instrument. UV dose was selected by measurement of cell viability, and ELISA results of DNA photoproduct. Optimum extracts condition of grape skin selected by DNA photoproduct content as well as at this condition; Bax/Bcl-2 ratio and cytochrome c gene expressions were evaluated by Western blotting.
RESULTS:
In this study, we confirmed that GSE protect against DNA damage-induced cell death. Condition for 80% EtOH for 24 h at 60 °C was suitable to extract for grape skin. Red grape skin (Zweigelt) was more effective than white grape skin (Niagara) in preventing (before irradiation) and repairing (after irradiation). UV-induced upregulation of Bax/Bcl-2 ratio and cytochrome c expression were reduced by GSE treatment.
CONCLUSIONS:
The study demonstrated a promising potential of GSEs in skin therapeutics application.
1Introduction
The amount of ultraviolet radiation (UVR) reaching the earth’s surface has increased, putting the health of living beings in danger, and affecting the quality of life on earth [1]. UVR is a type of electromagnetic radiation with wavelengths ranging 10–400 nm. As it is shorter than the violet part of sunlight, UV is invisible to the naked eye, but has a higher energy than visible light. Spectral distribution of sunlight is categorized into three wavelength bands: UVA (315–400 nm), UVB (280–315 nm), and UVC (200–280 nm). Among these, UVC has the highest energy and is extremely damaging to biological tissues [2]. UVC is unable to penetrate the earth’s atmosphere, and humans are mostly exposed to UVB and UVA, with the former possessing higher energy [3]. However, some medical treatments and industries use the UVC region, making humans susceptible to the detrimental effects of this radiation [4].
The skin and the eyes, have the greatest acute or chronic exposure to UVR, eventually resulting to different health impacts. Sunburn, erythema, epidermal hyperplasia, and inflammation occur as a consequence of short-term UVR exposure. Whereas, if the skin had long been exposed to UVR, photo-aging, immunosuppression, or carcinogenesis can occur [5]. On the other hand, UVR exposure can lead the eyes to contract dangerous cataracts and retinitis pigmentosa [6]. With humans’ increased risk of exposure and the known detrimental health effects of UVR, it is important to conduct studies on means to protect humans from UVR’s harmful effects.
UVA and UVB irradiation may give further skin damage due to destruction of vitamin A, which plays a significant role in maintaining skin elasticity and promoting skin renewal [7]. Moreover, UV from 245 to 290 nm can induce direct DNA lesions. Damaged DNA is responsible for oncogenic alterations, and plays an important role in triggering the action of cancers. Long time exposure to UVB or UVC can result to mutagenic DNA lesions through formation of dimeric photoproducts at pyrimidine sites, i.e. cyclobutane pyrimidine dimer (CPD) and (6-4) photoproduct (6-4 PP) [8]. CPD and 6-4 PP are formed between adjacent pyrimidine structures, such as the TC and CC residues of DNA. CPD is a 4-membered ring formed from the double bonds of C-5 and C-6 of the consecutive bases, while 6-4 PP arises from bond formation at 5’-C-6 and the 3’-C-4 [9, 10].
The changes in the DNA structure caused by UV can further lead to genetic mutations, which affect cell function and proliferation. These mutations may give rise to anomalies in cells, such as cancer. When cells are unable to repair the DNA damage, apoptosis occurs in order to prevent the acquisition of mutated genes to cells upon replication [11]. Apoptosis or programmed cell death is a distinct mode of cell destruction. It is a major regulatory mechanism in eliminating abundant and unwanted cells during embryonic development, growth, differentiation, and normal cell turnover. Apoptosis can occur through the induction of cytochrome c release from the mitochondria via activation of Bax protein (a Bcl-2 family member) [12, 13].
Plant extracts, containing large amounts of phenolic compounds, were reported to have beneficial antioxidant effects, and can potentially diminish the damage caused by UVR [14–17]. It was suggested that plants rich in phenolic compounds can modulate gene expression. In line with this, polyphenols were regarded as a feasible inhibitor affecting signal transduction pathways [13, 14, 16, 17].
Grape skins, waste products after fermentation of wine, are inexpensive sources of polyphenolic compounds (i.e., flavonoids), that have different nutritional and therapeutic benefits [18]. It has been reported that grape skin extracts (GSEs) play a positive role in interfering or reducing the progression of diseases and cancer [19, 20], in contrast grape seed extraction was found to have baleful effect, such as reduced effectiveness of oxaliplatin in colon cancer cell [21]. The role of GSE in the cellular apoptotic pathway is linked to the obstruction of anti-apoptotic molecules, as well as direct triggering of the pro-apoptotic signaling cascades [22, 23]. However, the effects of GSE in reducing DNA damage and apoptosis caused by UV irradiation in human skin cells have yet to be fully investigated.
This study aims to investigate the effectivity of GSE in diminishing UV-induced cytotoxicity in normal human epidermal keratinocytes (NHEK) cells through the measurement of cell viability, quantification of DNA photoproducts, and analysis of pro-apoptotic protein expressions. Treatment variables such as the variety of grapes, cell incubation time, extraction procedures, as well as treatment sequence, are taken into consideration in order to determine the optimum condition to ensure cell protection. This study starts with the confirmation of GSEs’ effectivity in reducing UV-induced damage, followed by the comparison among grape varieties. The inhibition of photoproduct formation was also explored in connection with the variation of extraction conditions and differences in amounts of obtained polyphenols. In addition, cellular pathways in which GSE may function as an important factor, was evaluated.
2Materials and methods
2.1Materials
Pooled “Neonatal Normal Human Epidermal Keratinocytes” (NHEK)-neo cells purchased from Lonza (00192907, Walkersville, MD, USA), from a neonatal single donor (male) sourced, all analyses were based on previous published studies, thus no ethical approval and patient consent are required. Keratinocyte Basal Medium (KBM) –GOLD, HEPES buffered saline (HEPES-BBS), trypsin/EDTA solution, and trypsin neutralizing solution (TNS), were all purchased from Lonza (Walkersville, MD, USA). Monoclonal antibodies against CPD and photoproduct (6-4 PP) were ordered from Cosmo-Bio Co. Ltd (Japan). Ribonuclease A (RNase), streptavidin-conjugated peroxidase, peroxidase-conjugated avidin, ethidium bromide, and o-phenylendiamine dihydrochloride (OPD) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Biotinylated goat anti-mouse immunoglobulin was purchased from Amersham Pharmacia Biotech (Buckinghamshire, England). DNA marker was from Promega (Madison, WI, USA). High pore PCR template preparation kit and proteinase K were purchased from Roche Diagnostics (Mannheim, Germany). Trypan blue staining solution (0.4%) was purchased from Bio-Rad (Hercules, CA, USA). ECL western blotting detection reagent was purchased from Amersham Pharmacia Biotech (Buckinghamshire, England). Other chemicals used were of analytical grade.
2.2Grape skin extract
The grape skins (Red grape skin Zweigelt; RGZ and white grape skin Niagara; WGN) underwent air-drying and blended with a blender (TSK-935J, EUPA, China) to make grape skin powder.
1) After that 10 g of grape skin powder was continuously mixed with 100 mL of methanol for 25 h at room temperature (25 °C). The grape skin broth was centrifuged (Kubota 2800, Kubota, Japan) at 3,000 rpm for 10 min and the supernatant was filtered using 0.45 μm pore filter paper (Asfil-CA, As one corporation, China) (for experiments in Figs. 2 and 3).
2) In optimization experiment, 1 g of grape skin powder was continuously mixed with 25 mL of methanol or ethanol for 0.5–25 h at a range of 25–60 °C (Table 1) (for experiments in Figs. 4 and 5). In Figs. 6 and 7, the samples were treated with 25 mL of 80% ethanol for 2 h/24 h at 25 °C (the optimum conditions in extraction by data of Figs. 4 and 5). Then the grape skin broth was centrifuged (Kubota 2800, Kubota, Japan) at 3,000 rpm for 15 min, and the supernatant was filtered using 0.45 μm pore filter paper (Asfil-CA, As one corporation, China).
Fig. 1
The vertical axis shows the required exposure dose at each wavelength to produce the same amount of CPD when UV irradiation of half-lethal dose at 250 nm produces CPD. The horizontal axis indicates the wavelength (nm). The reproducibility of this experiment has been confirmed in several experiments.
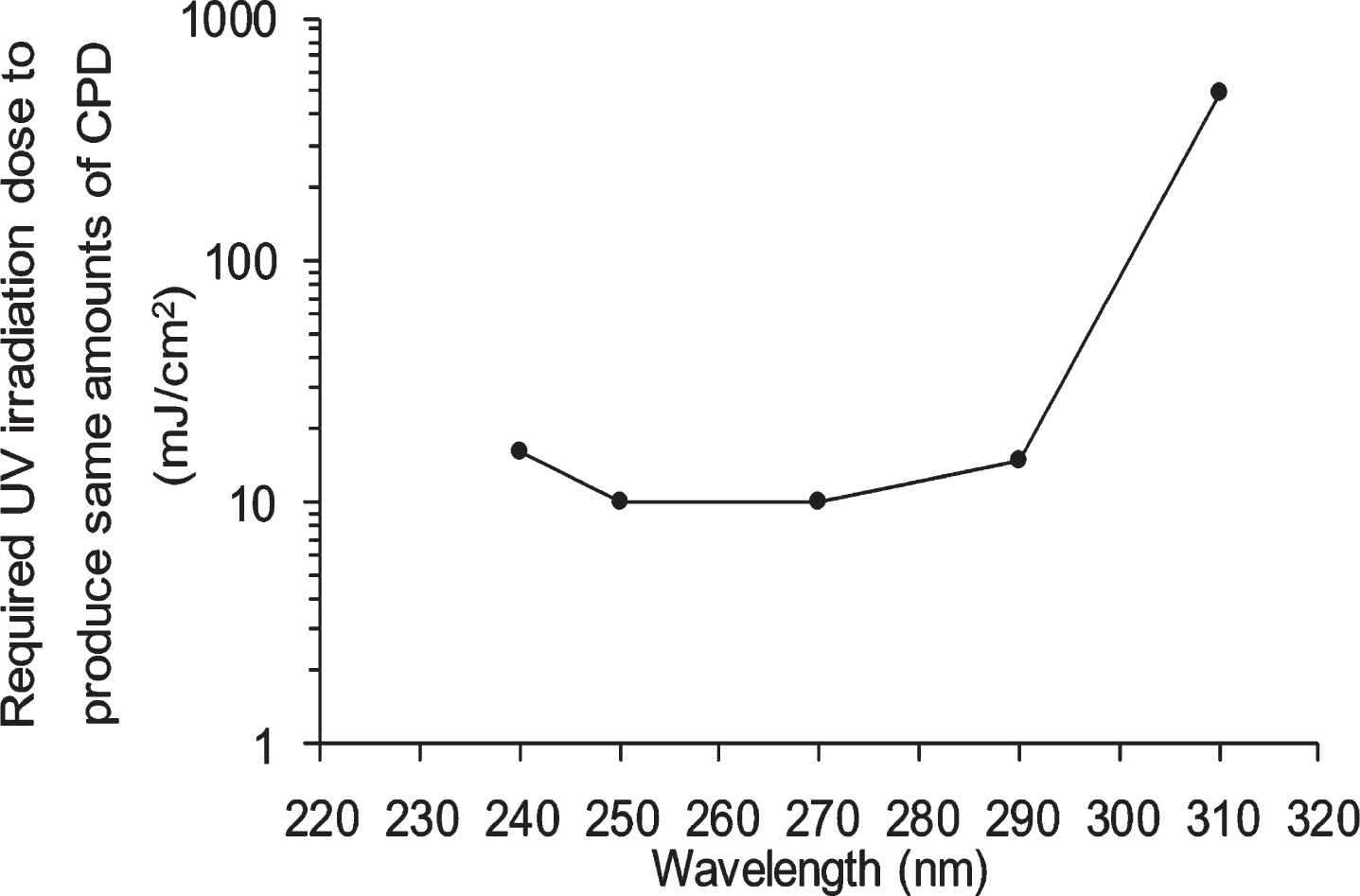
Fig. 2
Viability of NHEK cells treated with ethanol, white grape skin Niagara (WGN) extract and red grape skin Zweigelt (RGZ) extract, incubated for 1 h/24 h after UV irradiation at different wavelengths (250 nm, 270 nm, 290 nm, and 310 nm). “a”, “aa” and “aaa” denote significant change between 1 h and 24 h in each EtOH, WGN, and RGZ, and mean p < 0.05, p < 0.01, and p < 0.001, respectively. In control group treated without UV, ethanol was added instead of GSE. GSEs were obtained as described in method 1, section 2.2. Error bars indicate SEM (n = 3).
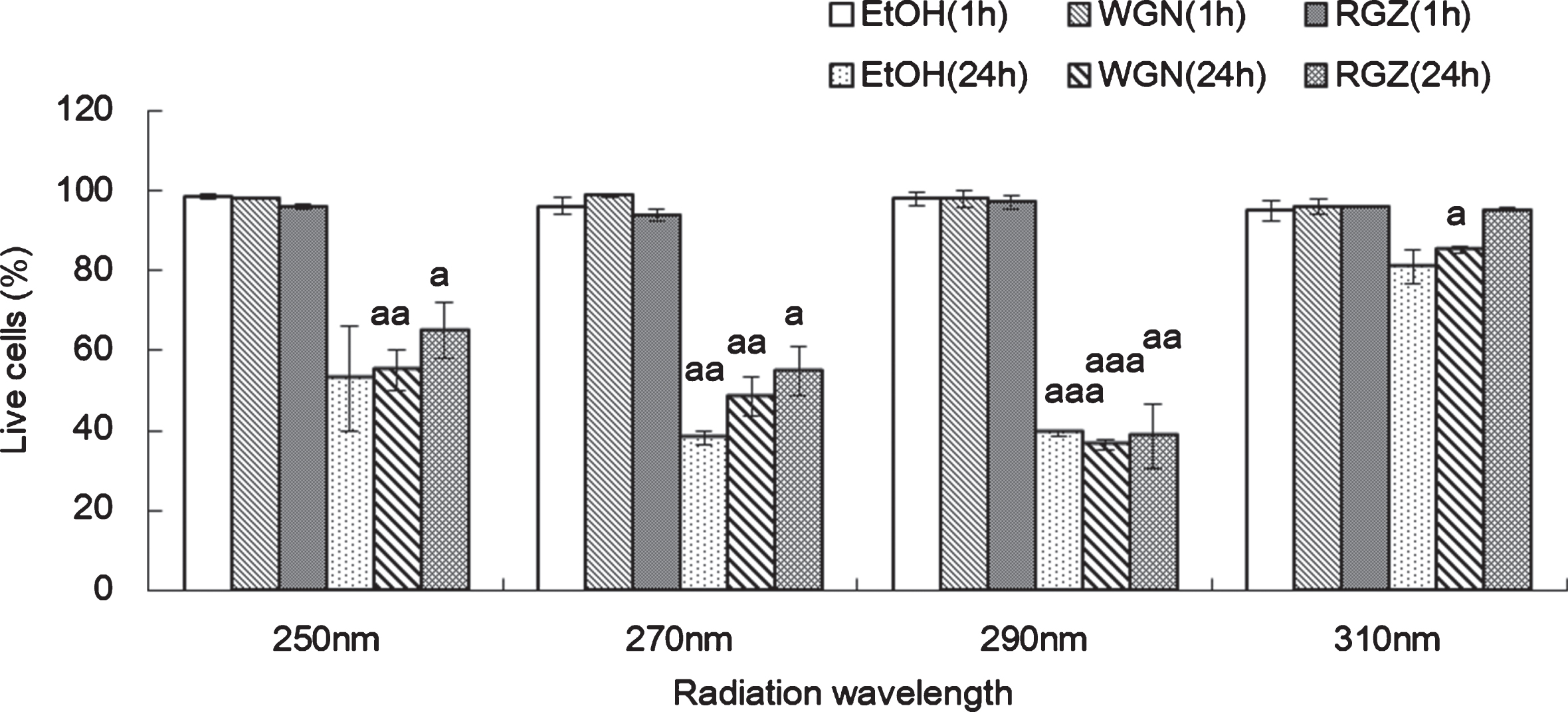
Fig. 3
CPD formation in NHEK cells treated with ethanol, WGN extract and RGZ extract 1 h (A) or 24 h (B) after UV irradiation at 4 different wavelengths (250 nm, 270 nm, 290 nm, and 310 nm). Asterisks denote significant difference *:p < 0.05, **:p < 0.01, ***:p < 0.001 compared to the NHEK cells treated with EtOH. “a” and “aaa” denote significant changes between WGN and RGN in each wavelength, and mean p < 0.05 and p < 0.001, respectively. In control group treated without UV, ethanol was added instead of GSEs. GSEs were obtained as described in method 1, section 2.2. Error bars indicate SEM (n = 3).
Table 1
Extraction conditions of grape skins after wine pressing
| No. | breed | solvent | Solvent concentration (% v/v) | extraction time (h) | extraction temperature (°C) |
| 0 | Zweigelt red | EtOH | 15 | 0.5 | 60 |
| 1 | Zweigelt red | EtOH | 80 | 0.5 | 60 |
| 2 | Zweigelt red | EtOH | 80 | 2 | 60 |
| 3 | Zweigelt red | EtOH | 80 | 24 | 25 |
| 4 | Zweigelt red | EtOH | 50 | 0.5 | 60 |
| 5 | Zweigelt red | EtOH | 50 | 24 | 25 |
| 6 | Zweigelt red | MeOH | 80 | 0.5 | 25 |
| 7 | Zweigelt red | MeOH | 80 | 24 | 25 |
| 8 | Zweigelt red | MeOH | 50 | 24 | 25 |
| 9 | Niagara white | EtOH | 80 | 0.5 | 60 |
| 10 | Niagara white | EtOH | 80 | 2 | 60 |
| 11 | Niagara white | EtOH | 80 | 24 | 25 |
| 12 | Niagara white | EtOH | 80 | 48 | 25 |
The filtered (both parts 1 and 2) was stored at –20 °C for future experiments. 1.5 mL of filtered solution was dried at room temperature (25 °C) with a SpeedVac Concentrator (Formingdale, NY, USA) and then mixed with 50 μL of ethanol, after that 2 μL of these broths were added in UV exposure experiment, and 2 μL of ethanol was added as control group. As results, extract materials from 6 mg (Figs. 2, 3) or 2.4 mg (Figs. 4–7) of grape skin (WGN or RGZ), were added in 35 mm culture dishes with 2 mL cell medium.
Fig. 4
CPD formation in NHEK cells 1 h after UV irradiation at 250 nm at different extraction conditions of RGZ. The numbers on the horizontal axis correspond to the numbers in Table 1. Asterisks denote significantly different values. *:p < 0.05, **:p < 0.01, ***:p < 0.001 as compared with NHEK cells treated with EtOH. In control group treated without UV, ethanol was added instead of GSEs. GSEs were obtained as described in method 2, section 2.2. Error bars indicate SEM (n = 3).
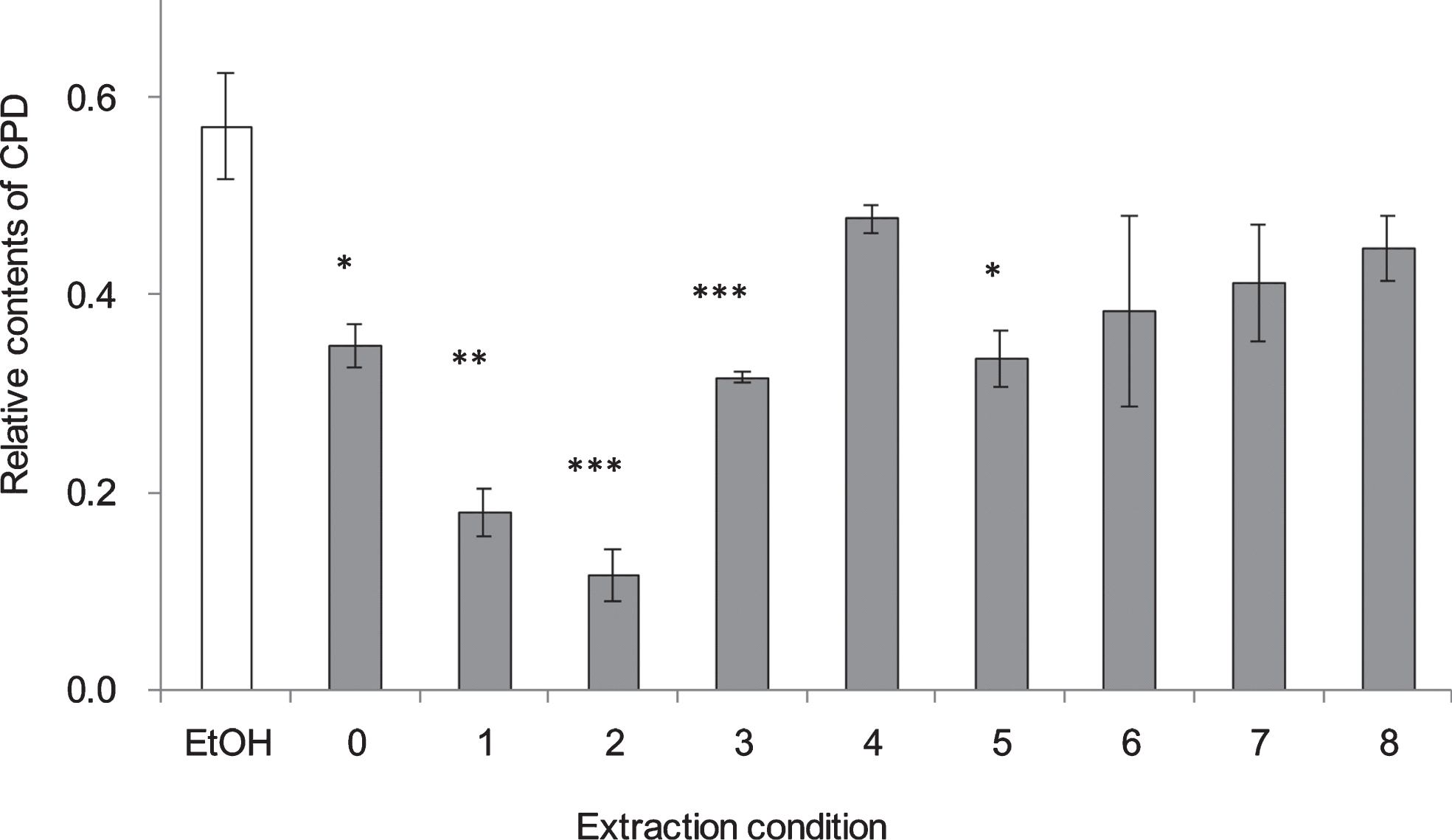
Fig. 5
CPD formation in NHEK cells 1 h after UV irradiation at 250 nm at 4 different extraction conditions of WGN. The numbers on the horizontal axis correspond to the numbers in Table 1. In control group treated without UV, ethanol was added instead of GSEs. GSEs were obtained as described in method 2, section 2.2. Error bars indicate SEM (n = 3).
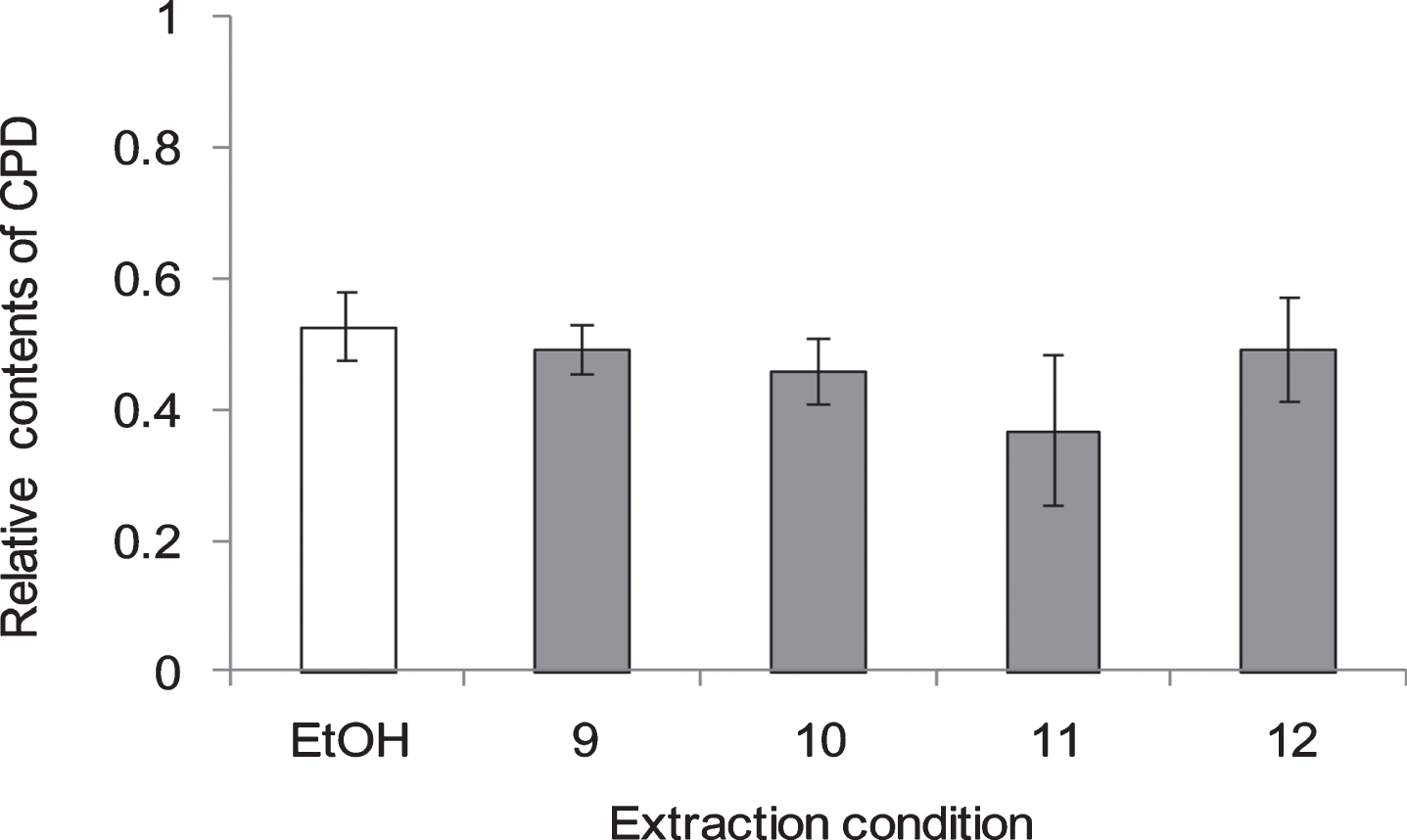
Fig. 6
CPD and 6-4 PP formation in NHEK cells treated with GSEs for 1 h before or after UV irradiation at 250 nm. Asterisk denotes significant difference *:p < 0.05, **:p < 0.01, ***:p < 0.001 as compared with the NHEK cells treated with EtOH, a denote significant change from WGN and RGN at p < 0.05, p < 0.01. In control group treated without UV, ethanol was added instead of GSEs. RGZ and WGN extraction condition was followed condition 2 and 11 of Table 1, respectively. Error bars indicate SEM (n = 3).

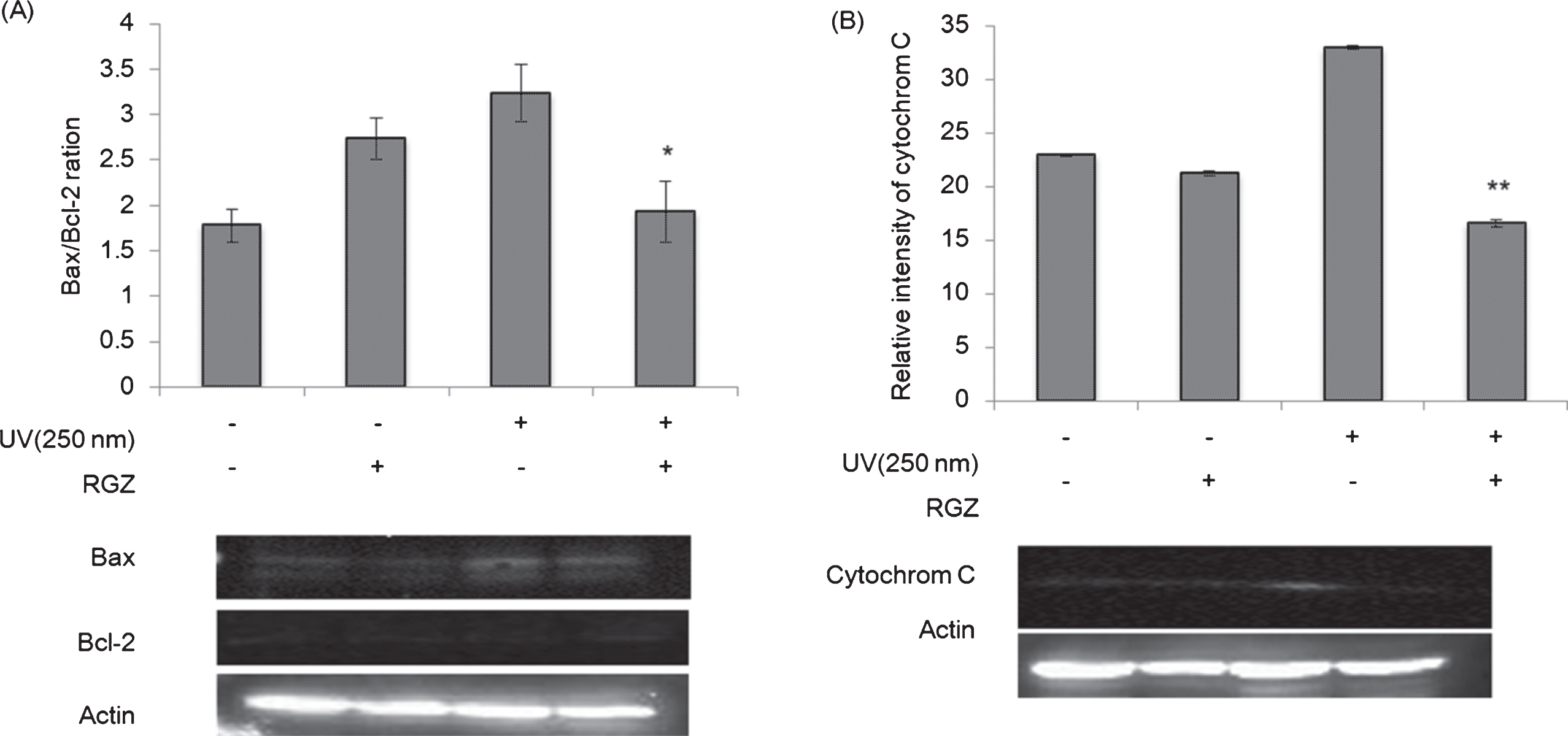
Fig. 7
Western blot analyses of Bax/Bcl-2 ratio (A) and cytochrome C (B) in NHEK cells treated with and without RGZ under the UV irradiation (250 nm). Asterisk denotes significant difference *:p < 0.05, **:p < 0.01 as compared with the NHEK cells treated with UV-only treated group. RGZ extraction condition was followed condition 2 of Table 1. Error bars indicate SEM (n = 3).
2.3Cell culture and GSE treatment
NHEK cells were maintained in KBM-Gold medium, and incubated (Direct heat incubator, Astec, Japan) at 37 °C with 5% CO2 in a 25 cm2 flask, until the desired confluency. Before treated with UV irradiation, NHEK cells (3–4 passage) at a density of 20,000 cells per well, were cultured in 35 mm culture dishes with 2 mL KBM-Gold medium, and placed in 5% CO2 incubator (Direct heat incubator, Astec, Japan). The cells for transfer were washed with 5 mL of HEPES-PBS, and 2 mL of trypsin/EDTA solution was added. When all of the cells were floating, 5 mL of TNS was added to stop the trypsin reaction. Cells were then transferred to a 15 mL of sterilized polyethylene tube, and centrifuged (Kubota 2800, Kubota, Japan) for 3 min under 1500 rpm at 4 °C. Finally, cells were cultured in 35 mm culture dishes with 2 mL KBM-Gold medium, and placed in CO2 incubator (Direct heat incubator, Astec, Japan) as prepared to UV expose.
Two μL of the GSE was added (before or after expose to UV), and the cells were incubated (Direct heat incubator, Astec, Japan) for 1 h or 24 h. In control group, 2 μL of ethanol was given because ethanol cannot affect cell condition [24].
2.4UV treatment
A xenon-lamp light source (MAX–301, Asahi Spectra, Tokyo, Japan), equipped with bandpass filters capable of isolating specific wavelength regions, was selected as the UV source. Prior to UV exposure, irradiance was measured at the position of the target cells, using a calibrated radiometer (IL 1400A, International Light Technologies, Peabody, USA) connected to a silicon photodiode detector (SEL033, International Light Technologies). Irradiation intensities were chosen on the basis of the methodology as described by Shirai [24]. Exposure time was calculated from dividing irradiation intensity by irradiance (Equation 1).
Wherein, irradiance at the target position (mW/cm2) was determined, by getting the difference between UV readings after and before exposure (UV after –UV before).
NHEK culture medium is able to absorb UV irradiation, which can affect the amount of irradiation that is being treated to the cells [25]. Therefore, the KBM-Gold medium was modified through addition of 2 mL PBS. The original culture medium was first discarded, and the cells were washed twice with 1 mL of PBS, followed by the addition of the modified KBM-gold medium into the culture. Then, the culture dishes were placed on a narrow band with a width of approximately 10 nm, and the cells were irradiated at different wavelengths of UV.
NHEK cells in 35 mm culture dishes with 2 mL of PBS were exposed to UV radiation at different wavelengths from 240 to 310 nm, with several dose to determine same amounts of CPD formation in each wavelength. The based CPD amounts were defined, as those produced by LD50 dose at 250 nm (10 mJ/cm2). Upon the completion of irradiation treatment, PBS was discarded and replaced with KBM-Gold, and then the NHEK cells were cultured for 1 h. The expose dose required to normalize DNA damage, was determined by UV irradiation dose, which produced the same amounts of CPD in each wavelength (Fig. 1). After each dose was determined, the effect of GSE was examined.
2.5Cell viability
Cell viability was determined using trypan blue exclusion assay at 1 and 24 h after the UV irradiation. After incubation, cells were harvested, washed with PBS, and stained in 0.2% trypan blue solution. Total cells and trypan blue staining cells, were counted by an automated cell counter (Bio-Rad, Hercules, CA, USA). Cell viability was expressed as a percentage of live cells against the total cell number. Each experiment was repeated for at least three times to ensure reproducibility.
2.6Isolation of genomic DNA from NHEK cells
Genomic DNA was isolated using a high pure PCR template preparation kit (Roche; Mannheim, Germany). After irradiation, NHEK cells were harvested using typsin/EDTA solution. After two times washing and centrifugation (Kubota 2800, Kubota, Japan), the supernatant was discarded, and 200 μL PBS was added to loosen the cells. Then, 200 μL of binding buffer (10 mM Tris-HCl pH 4.4, containing 6 M guanidine-HCl, 10 mM urea and 20% Triton X-100) and 40 μL of proteinase K (20 μg/mL) was added, and the solution was incubated (Direct heat incubator, Astec, Japan) at 70 °C for 10 min. After addition of 100 μL isopropanol, the solution was transferred into a high pure filter tube attached to a collection tube, which was centrifuged (Kubota 2800, Kubota, Japan) at 12,000 rpm for 1 min. The filter tube was then attached to a new collection tube, and 500 μL inhibitor removal buffer (5 M guanidine-HCl and 20 mM Tris-HCl) was added, followed by centrifugation (Kubota 2800, Kubota, Japan) at the same condition. The obtained precipitate was washed and centrifuged (Kubota 2800, Kubota, Japan) with 500 μL washing buffer (2 mM Tris-HCl, pH 7.5 containing 20 mM NaCl) twice. The DNA solution was obtained through addition of pre-warmed 200 μL elution buffer to the filter tube, followed by centrifugation (Kubota 2800, Kubota, Japan) at 12,000 rpm for 1 min. Afterwards, 2 μL of 500 μg/mL RNase was added and the solution was incubated (Direct heat incubator, Astec, Japan) for 15 min at 37 °C. Finally, 500 μL of ethanol and 20 μL of 3 M NaOAc buffer (pH 4.5) were added for DNA precipitation, and the solution was allowed to stand overnight at –20 °C.
2.7ELISA analysis for CPD and 6-4 PP formation
2.7.1DNA concentration adjustment for ELISA
Genomic DNA obtained from NHEK cells was centrifuged (Kubota 2800, Kubota, Japan) at 15,000 rpm for 7 min at 4 °C. To remove the insoluble salts, 500 μL of 70% ethanol was added. Afterwards, the solution was centrifuged (Kubota 2800, Kubota, Japan) at 15,000 rpm for 3 min at 4 °C to obtain the precipitated DNA, which was then dried using a SpeedVac Concentrator Savant SVC 100 H (Instruments, Formingdale, NY, USA). Dried DNA was dissolved in 50 μL 1×TBE and the absorbance of the solution was measured at 280 nm with a Personal Spectrum Monitor (Gene Quant Pro, GE, UK). In order to equalize the DNA concentration in all samples, 1×TBE was added up to a concentration of 1 μg DNA/μL.
2.7.2ELISA of CPD and 6-4 PP in NHEK cells
The contents of CPD and 6-4 PP in the extracted genomic DNA were measured by ELISA. 100 μL of each sample containing 4 μL of DNA was placed in titer plate wells and incubated for 1 h at 37 °C. After incubation, the titer plate was washed three times with 0.03% of Tween 20 and 0.9% NaCl in 40 mM Tris-HCl buffer, then blocked with 100 μL of 2% blocking reagent for 30 min at 37 °C. The first antibody for CPD or 6-4 PP (1 : 1000 diluted with tween buffer), was immobilized with antigens in the samples for 1 h at 37 °C. Then, the plate was washed again with tween buffer three times. The second antibody, a biotinylated anti-mouse immunoglobulin (1 : 500 diluted with 0.03% Tween 20 in 40 mM Tris-HCl buffer and 0.9% NaCl) was added, and the plate was incubated (Direct heat incubator, Astec, Japan) for 1 h at 37 °C. The plate was re-washed with tween buffer three times. After addition of streptavidin-conjugated peroxidase, (1 : 400 diluted with tween buffer) the plate was incubated (Direct heat incubator, Astec, Japan) for 30 min at 37 °C. After washing, 0.1% OPD in 50 mM phosphate-citrate buffer pH 5.0, containing 0.03% sodium perborate, was added into the cells and 5–10 min later, HCl was added to stop the enzymatic reaction. Absorbance at 495 nm was measured with a Microplate Reader model 450 (Bio-Rad).
2.8Western blot analysis for Bax, Bcl-2 and cytochrome c
After exposure to UV, NHEK cells were cultured with or without GSE for 1 h. Then, cells were washed with PBS twice and harvested. The obtained cells were centrifuged (Kubota 2800, Kubota, Japan) for 1 min at 1,500 rpm, 4 °C. For cytochrome c measurement, after discarding the supernatant, cytochrome c release Apoptosis Assay kit (Calbiochem, Darmstadt, Germany) was used as extraction buffer. For the measurement of Bax (BIS, bs-0127 R, BIOSS), Bcl-2 (Cat#PC68, Oncogene) and beta-actin (GTX 109639, GeneTEX) the cell lysis buffer used, was constituted of 2 mM HEPES, 100 mM NaCl, 10 mM EGTA, 1 mM, PMSF, 0.1 μM MPMSF, 1 mM Na2MgO4, 5 mM 2-glycerophosphoric acid, 10 μM MgCl2, 2 mM DTT, 50 μM NaF and 1% Triton X-100.
The cells were soaked in 70 μL of the respective buffer for the desired protein. Physical extraction of proteins was done through two-time freeze-thawing and sonication cycles. The concentration of proteins in cells was determined spectrophotometrically, using a protein assay dye reagent. 25 μg of proteins from each sample was separated by 12.5% polyacrylamide gel electrophoresis using electrophoresis system (Mupid-ex, Advance, Tokyo, Japan). Semi-dry western blot transfer method was employed in transferring the proteins to a nitrocellulose membrane, which was then incubated overnight in 5% blocking buffer at 4 °C. Incubation (Direct heat incubator, Astec, Japan) with the first antibody was done for 1 h at 37 °C, followed by washing with tween buffer thrice. The membrane was incubated (Direct heat incubator, Astec, Japan) with secondary antibody at 37 °C for 1 h, and the membrane was washed again for three times. Protein band visualization was done using ChemiDoc XRS (Bio-Rad, USA), and protein densities in different lanes were analyzed using Image J.
2.9Analysis of polyphenol content in GSEs
The grape skin powder samples, obtained as described in step 2.2, were dissolved in 100 mL of methanol. Methanol extracted from grape skins, was dried and dissolved again in 100 μL of methanol, and examined by ESI-MS and HPLC, in at least three independent experiments. ESI-MS was performed with a JMS-LCmate (JEOL, Tokyo, Japan) in positive ionization mode (ESI+) with a methanol carrier solution at a 0.5 mL/min flow rate and injection volume of 2 μL.
HPLC was performed with an MD-1510 and a reverse-phase CRESTPAK C18 S column (150×4.6 mm; Jasco, Tokyo, Japan). Data were analyzed by DP-L1500 W (Jasco). The mobile phase consisted of acetonitrile and H2O in a 15 : 85 ratio (v/v). The mobile phase was filtered through a 0.2-μm membrane, and degassed by aspirator before analysis. A 20-μL sample was separated by the column at 1.0 mL/min flow rate, and authentic standards of resveratrol and anthocyanins (Sigma –Aldrich) were used. All experiments were performed at ambient temperature.
Contents of total polyphenol were determined by Folin-Ciocalteu method, using Folin’s reagent (detecting absorbance at 765 nm) and utra high performance liquid chromatography (Shimazu Nexera-iMT), respectively.
2.10Statistical analysis
Each value is expressed as the mean±Standard Error of Mean (SEM). Statistical analyses were performed by one-way analysis of variance (ANOVA), followed by the Fisher’s test.
3Results
3.1UV dose selection
To normalize DNA damage in NHEK cells, the dose of UV radiation that produced the same amount of CPD, was determined (Fig. 1). The curve appeared a broad minimum in the wavelength of approximate 250–290 nm, it means that UV in this extend is most risky. The damage levels of DNA in the NHEK cells caused by UV irradiation with 10 mJ/cm2 at 270 nm, 15 mJ/cm2 at 290 nm and 500 mJ/cm2 at 310 nm were same as that with 10 mJ/cm2 at 250 nm (Fig. 1). These four conditions: 10 mJ/cm2 at 250 nm, 10 mJ/cm2 at 270 nm, 15 mJ/cm2 at 290 nm, and 500 mJ/cm2 at 310 nm, were employed in subsequent experiments to clarify the effect of GSE on DNA damage caused by UV irradiation.
3.2Evaluation of viability of cells exposed to GSEs
NHEK cells were exposed to 4 different wavelengths of UV radiation with the following intensities: 10 mJ/cm2 at 250 nm and 270 nm, 15 mJ/cm2 at 290 nm, and 500 mJ/cm2 at 310 nm. To examine whether GSE have positive effects on protecting cells from UV irradiation-induced toxicity, the cell viability was measured by trypan blue staining at 1 and 24 h after the UV irradiation. It must be noted that, ethanol was used as control and poses no toxicity on NHEK cells (data not shown). As shown in Fig. 2, there is no significant difference in viabilities of cells cultured with and without GSE for 1 h after 4 different dose (250 nm. 270 nm, 290 nm, and 310 nm) of UV irradiation, meaning that, 1 h incubation post-UV irradiation at all wavelengths was not sufficient to induce significant cell viability alteration.
At 24 h after 4 different dose of irradiation, although there are no considerable differences among cells exposed to ethanol, RGZ, and WGN, there was an observable tendency that GSEs demonstrated protective behavior on cells as compared with only ethanol. In detail, the viabilities of cells treated with ethanol and exposed to UV irradiation at 250 nm and 270 nm, were 53% and 38%, respectively. On the other hand, WGN extracts increased cell viability in both wavelengths by 2% and 10%, respectively. Furthermore, RGZ-treated cells had an increase in viability by 12% at 250 nm and 17% at 270 nm, as compared to ethanol-only treated cells. It was observed that cells treated with RGZ extract exhibited higher viability than WGN-treated cells. In addition, the highest viabilities of NHEK cells at 24 h after UV irradiation (across all treatment types) were attained at 310 nm and the lowest were found at 290 nm.
After treatment with each irradiation of UV (250 nm. 270 nm, 290 nm, and 310 nm) the significant decline of cell viability in incubated cells for 24 h as compared to 1 h indicates that a longer incubation time can induce the observable toxicities of shortwave irradiation.
3.3CPD measured by ELISA analyses
3.3.1UV irradiation in four different wavelengths
One hour cultivation post-UV irradiation, the amounts of synthesized CPD in cells treated with GSEs were reduced dramatically, in comparison to the ethanol-treated group; indicating that both RGZ and WGN exhibited protective capabilities against the UV irradiation-triggered DNA alteration after treatment with 4 different does irradiation of UV (250 nm. 270 nm, 290 nm, and 310 nm). Comparing RGZ and WGN-treated cells, the former contained a lower amount of generated CPD than the latter in 250 nm and 270 nm wavelength, thus, it can be inferred that at 1 h post-UV irradiation (250 and 270 nm) RGZ possessed a stronger inhibition capability on the generation of the photoproduct CPD (Fig. 3 (A)).
3.3.2Twenty four hour after UV irradiation at 290 nm and 310 nm
24 h cultivation after UV irradiation at 250 nm and 270 nm, the content of CPD generated in the NHEK cells treated with either RGZ or WGN extracts, were lesser than ethanol group. Comparing to the case of 1 h treatment, especially at 270 nm, the amount of generated CPD was decreased by more than 50% in RGZ extracts. With respect to cell viability data (Fig. 2, 250 nm and 270 nm), it is suggested that damaged cells survive through DNA repair action, which may be induced by GSE. Therefore, a relationship can be deduced in which culture time after the UV treatment, is directly proportional with repair rate; wherein, a longer incubation time after irradiation would allow cells to repair the UV-damaged DNA and remove the photoproducts like CPD (Fig. 3 (B), 250 nm and 270 nm).
In cells incubated for 24 h after UV irradiation (Fig. 3 (B)), the production of CPD in NHEK cells treated with WGN extract at 290 nm, was decreased as compared to cells treated with ethanol. However, the cells treated with RGZ shows an opposite trend, wherein a slight increase in the CPD content was observed. Unexpectedly, at 290 nm and 310 nm UV irradiation, CPD production reached its highest in cells treated with RGZ. In contrast to data at 250 nm and 270 nm, RGZ showed minimal effect on reducing the CPD production at both 290 nm and 310 nm. This finding indicates that the variety of the grape skin is not the only factor affecting DNA damage repair, but the wavelength of the irradiation can also play a crucial role. UV waves with a relatively low frequency (high in wavelength) can hinder the capability and efficiency of GSEs on DNA repair.
3.4Condition of grape skin extraction
To evaluate the extraction method efficiency of RGZ and WGN in reducing CPD, several extraction variables were examined (Table 1), such as the solvent type and concentration, together with the extraction period and temperature. From the results shown on Fig. 4, it was determined that the most efficient extraction condition in lessening CPD formation was 80% ethanol, for 2 h at 60 °C (Condition no. 2). Unexpectedly, there was no remarkable changes among extract conditions of WGN in reduce CPD formation. On the other side, no. 11 : 80% of ethanol, 24 h at room temperature, showed the lowest CPD formation when compare with other extract conditions, which use for extracts condition in follow study.
3.5Resveratrol and polyphenol contents in GSEs
The amount of polyphenols extracted per volume of GSEs is displayed in Table 2. It was found that the anthocyanin content of RGZ at different extraction conditions, were found to be relatively higher than that of WGN. On the other hand, resveratrol content in WGN were higher than that in RGZ. Total polyphenol amount in the red grape (Zweigelt), however, was almost twice as much as it was in the white variety (Niagara). From the results, the difference between total polyphenols and anthocyanins may be the reason why RGZ has better inhibitory ability on CPD production than WGN.
Table 2
Amounts of polyphenols per volume of GSE
| Extraction no. | Sample and extraction method | Total anthocyanin (mg/100 mL) | Resveratrol (mg/100 mL) | Total polyphenol (mg/100 mL) |
| 0 | Zweigelt red (15% EtOH, 30 min, 60 °C) | 5.9 | 0.11 | 42.6 |
| 1 | Zweigelt red (80% EtOH, 30 min, 60 °C) | 8.3 | 0.45 | 46.6 |
| 2 | Zweigelt red (80% EtOH, 2 h, 60 °C) | 8.5 | 0.46 | 51.4 |
| 3 | Zweigelt red (80% EtOH, 24 h, 25 °C) | 7.5 | 0.43 | 41.7 |
| 5 | Zweigelt red (50% EtOH, 24 h, 25 °C) | 9.4 | 0.40 | 61.9 |
| 7 | Zweigelt red (80% MeOH, 24 h, 25 °C | 8.3 | 0.43 | 43.1 |
| 10 | Niagara white (80% EtOH, 2 h, 60 °C) | 0.2 | 0.80 | 33.1 |
| 11 | Niagara white (80% EtOH, 24 h, 25 °C) | 0.1 | 0.70 | 24.7 |
The values in Table 2 show the average of two measurements in each sample. If the values were significantly different, a third measurement was performed, and the two closest values were averaged.
3.6Effect of treatment sequence of the GSEs on CPD and 6-4 PP production
To determine the most effective treatment sequence in suppressing the formation of CPD and 6-4 photoproduct from the cells, RGZ or WGN was added 1 h before or after treatment with 250 nm UV irradiation, and the photoproducts were measured using ELISA. As shown in Fig. 6(A), with the addition of RGZ 1 h prior to the irradiation, formation of CPD declined more than 80% compared to that of the ethanol group. WGN-treated cells had approximately 50% decrease on the CPD formation, compared to ethanol group. RGZ treatment 1 h after UV irradiation, led to common (but lower than before) degree of reduction on the CPD formation (50%), whilst WGN treatment had similar significant reduction compared to ethanol group (Fig. 6(B)).
It is worth noting that under both treatment conditions, the GSEs had a similar trend (Fig. 6 (C) and (D)). A significant difference in 6-4 PP production was found between the cells treated with GSEs and ethanol, 1 h before and after UV irradiation. Compared to ethanol group, nearly 50% and 80% reduction (before and after) on the 6-4 PP content was obtained from WGN and RGZ treatment, respectively. According to both CPD and 6-4 PP data, it can be inferred that the protective capability of RGZ is superior compared to that of WGN. Along with this, applying GSE to the cells 1 h before UV irradiation is more effective and sufficient, than treatment after the UV irradiation.
3.7GSEs defend UV-irradiated keratinocytes via inhibiting pro-apoptosis gene expression
To clarify the cellular mechanism effects of GSEs in UV-irradiated NHEK cells, we measured the expressions of pro-apoptosis factors Bcl-2, Bax and cytochrome c using western blot analyses. Although UV irradiation enhanced Bax/Bcl-2 ratio and cytochrome c contents, addition of RGZ suppressed these factors (Fig. 7). This implies that GSEs inhibited UV-induced apoptosis via mitochondrial pathway.
4Discussion
The efficiency of grape extracts have led to the growing acceptance of these natural products as a potential source of cancer chemo-preventive drugs [26]. Recently, significant development has been made in prostate cancer cells. Wherein, studies found that grape extracts have capabilities in restraining the cancer cells’ growth, and equal effectivity was seen in skin cancer cells [27]. A.M. Engelbrecht was able to conclude that pro-anthocyanidin from grape seed extract can exert its advantageous influence through enhancement of apoptosis in colon cancer cells [28]. However, the effects of GSEs in damaged normal human cells, have not been thoroughly investigated.
Our study evaluated the effectivity of GSEs on NHEK cell survival after exposure to various UV wavelengths, with regard to the sequence of treatment, incubation time, and type of GSE. Suffering same degree of DNA damage from UV light (Fig. 1), cell viability of NHEK cells cultured 24 h after 250, 270, and 290 nm UVR treatment, significantly decreased compared to the cells incubated for 1 h (Fig. 2), which was also found early in PC12 cell line [29]. An abundance of studies regarded CPD and 6-4 PP as measures to evaluate the UVR-induced DNA damage on cells [30–32]. It can be seen that, both cell death and DNA damage were observed in the cells incubated 24 h only after UVR (Figs. 2 and 3 (B): 250, 270, and 290 nm), which indicated that UV induced-DNA damage eventually led to cell death as incubation time extended from 1 h to 24 h. From this, a possible connection can be made between UVR-induced cell death and the incubation time length, which was explored through the evaluation of photoproduct removal as an apoptosis preventive measure. Evidences showed cell apoptosis leading to cell death started at 8 h incubation time after UVR [30]. This further implies that, at longer incubation time although DNA photoproduct amounts have declined, the repair process was not enough to ensure viability, and the cells proceeded to apoptosis. In this study, we provided more documentation that longer wavelengths, such as 270 nm and 290 nm, are in agreement with the results of the aforementioned study [30]. On the other hand, unlike CPD formation which was found significant decreased after expose to 310 nm of UV in incubate time (1 h and 24 h), the cell viability was no remarkable changed (Figs. 2 and 3), it suggest that for longer wavelengths, such as 310 nm, a longer exposure time or stronger dose of UV radiation is required in order to induce observable cell death caused by DNA damage [33].
The benefits of GSEs at each UV exposure conditions (Fig. 3) have provided sufficient evidence for decreasing DNA damage, which served as basis for continued research. CPD formation of cell with GSE added, had a drastic drop, and especially was observed at 250 nm; thus that wavelength of UV radiation was used for further experimentation.
To determine whether the GSE’s effect on UV-induced DNA damage is related to its extract methods or not, extract conditions were assessed based on CPD amounts. Continuous stirring of RGZ in 80% EtOH for 2 h at 60 °C, and WGN extraction in 80% EtOH for 24 h at RT, were found to be the most effective in preventing CPD formation (Figs. 4 and 5).
6-4 photoproduct is an essential determinant of the lethal and mutagenic effects of UV irradiation on biological systems [9]. The removal of this lesion appears to correlate closely with the early DNA repair responses of mammalian cells, including DNA incision events, repair synthesis, and removal of replication blocks [30]. The processing of 6-4 photoproduct and CPD appears to be enzymatically coupled in most mammalian cell lines examined [30]. In addition, the differences in the processing of the two photoproduct in some cell lines of human and rodent origin suggests that, in the case of mammalian cells different pathways in repairing 6-4 photoproduct and CPD may occur. Moreover, an earlier study reported that although CPD is the major photoproduct formed in the first 4 h after UV irradiation, 6-4 PP received priority in the repair process [34]. Taking into consideration the findings of the aforementioned study along with our data, which showed 6-4 PP amounts were considerably declined as compared to decrement in CPD, especially in case of RGZ treatment (Fig. 6), we can say that GSEs enable a DNA repair pathway that prioritizes the repair of 6-4 PP over CPD. Through the results (Fig. 6) and our methodology of washing out GSEs, we can attribute the photoproduct inhibition to the interaction of GSE chemicals to cells. CPD as the major products of DNA damage caused by UV irradiation, treating NHEK cells with GSE prior to UV exposure resulted to substantial decrement in CPD, as compared to treatment after UV irradiation. It indicate that the GSE effect was more preventive (1 h before irradiation) than repairing (1 h before irradiation). The trends derived from the data in Fig. 6, were RGZ > WGN, and GSE addition pre->post-UV irradiation, in terms of DNA photoproduct reduction efficiency. The exact mechanisms of inhibition of DNA photoproduct formation by GSEs, however, warrants further investigation.
From the experiment, the most interesting finding was that there was higher CPD and 6-4 PP levels on WGN-treated cells as compared to RGZ-treated ones, but are still considerably lower than the ethanol group. Numerous studies have attempted to explain the anticancer activity of grape extracts by focusing on resveratrol, one of the most famous compounds found in grape skin and seeds [23, 25, 35]. We first assumed that the decrease in CPD production was due to the amounts of resveratrol in each GSE. However, the resveratrol amount is not proportional with the GSEs’ capability to inhibit CPD production with RGZ, which having lesser resveratrol but higher CPD inhibition than WGN (Figs. 5 and 6, and Table 2). Earlier studies stated that anthocyanins were not found in white grapes [36, 37], which may support the difference in photoproduct amounts. On the other hand, anthocyanins were also found in white grape skin extraction (Table 2), which is different from such statement, but consistent with the study reported recently [38], it is consistent with our finding as seen in Table 2. Therefore, it can be said that the inhibition of photoproduct formation is not attributed to resveratrol nor anthocyanin alone, but to a variety of compounds, such as quercetin and pro-anthocyanidins. Such compounds have been found in grape skin through different extraction conditions, and have been implicated in possessing anti-cancer activity [25].
Reactive oxygen species (ROS) induced by UV irradiation, is one of the important mechanisms of apoptosis, activated by stimulation via mitochondrion. Bax/Bcl-2 ratio is an important standard in evaluating the cells going through survival or death in multicellular organisms. Upregulation of this ratio results to mitochondrial membrane permeability increase, followed by cytochrome c release, which then activates an array of related pro-apoptosis proteins leading to the induction of apoptosis [39]. Our data suggest that GSEs hinders induction of apoptosis in UV-treated NHEK cells by increasing Bcl-2 expression and decreasing Bax expression, further prohibiting cytochrome c release (Fig. 8). Of course, the possibility that ROS played an important role in this process could not be excluded.
GSEs contain a unique mix of polyphenolic compounds that possess beneficial properties such as anti-oxidant, anti-inflammatory, anti-cancer, anti-microbial, and anti-viral [18]. With these properties, GSEs have a higher potential in preventing the formation of UV-induced DNA photoproducts and cell apoptosis, as compared to using only one compound. Overall, effectivity of GSEs in protecting cells against UV-induced damage, is dependent on factors such as the culture time after expose to UV, variety of the grape, extraction conditions, and sequence of treatment (with respect to irradiation). From the study, it was found that red grape skin (RGZ) is more effective as compared to white grape skin (WGN); wherein, continuous stirring of RGZ with 80% ethanol for 24 h at 60 °C, is the best extraction method; and extracts should be added to the cells prior to UV irradiation. The study bears the potential in finding methods to prevent the adverse effects of UV radiation on the human skin through grape skin extraction. In which, cost-effective means of extraction can also aid in lessening waste from industries like winemaking, while producing natural products that can pave way in finding medical treatments.
Funding
The authors report no funding
Conflict of interest
The authors have no conflict of interest to report
Acknowledgment
The research was supported by grants from Hokkaido Wine Co., Ltd. through Hokkaido University. The authors declare that there is no conflict of interest. The authors extend their appreciation to Ms. Miyako Komori, Dr. Yongkun Sun, Dr. Mostafizur Rahman and Dr. Rachael U. Lopez for their advice and assistance in developing this study.
References
[1] | D’Orazio J , Jarrett S , Amaro-Ortiz A , Scott T . UV radiation and the skin. Int J Mol Sci. (2013) ;14: (6):12222–48. doi: 10.3390/ijms140612222 |
[2] | Ridley AJ , Whiteside JR , McMillan TJ , Allinson SL . Cellular and sub-cellular responses to UVA in relation to carcinogenesis. International Journal of Radiation Biology. (2009) ;85: (3):177–95. doi: 10.1080/09553000902740150 |
[3] | Tornaletti S , Pfeifer GP . UV damage and repair mechanisms in mammalian cells. Bioessays. (1996) ;18: (3):221–8. doi: 10.1002/bies.950180309 |
[4] | Nussbaum EL , Flett H , Hitzig SL , McGillivray C , Leber D , Morris H , et al Ultraviolet-C irradiation in the management of pressure ulcers in people with spinal cord injury: a randomized, placebo-controlled trial. Arch Phys Med Rehabil. (2013) ;94: (4):650–9. doi: 10.1016/j.apmr.2012.12.003 |
[5] | Beissert S , Granstein RD . UV-induced cutaneous photobiology. Critical reviews in biochemistry and molecular biology. (1996) ;31: (5-6):381–404. doi: 10.3109/10409239609108723 |
[6] | Pallela R , Na-Young Y , Kim S-K . Anti-photoaging and photoprotective compounds derived from marine organisms. Marine drugs. (2010) ;8: (4):1189–202. doi: 10.3390/md8041189 |
[7] | Törmä H , Berne B , Vahlquist A . UV irradiation and topical vitamin A modulate retinol esterification in hairless mouse epidermis. Acta dermato-venereologica. (1988) ;68: (4):291–9. doi: |
[8] | Rastogi RP , Kumar A , Tyagi MB , Sinha RP . Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. Journal of nucleic acids. (2010) ;2010: . doi: 10.4061/2010/592980 |
[9] | Mitchell DL , Nairn RS . The biology of the (6-4) photoproduct. Photochemistry and photobiology. (1989) ;49: (6):805–19. doi: 10.1111/j.1751-1097.1989.tb05578.x |
[10] | Setlow R . Cyclobutane-type pyrimidine dimers in polynucleotides. Science (New York, N.Y.). (1966) ;153: (3734):379–86. doi: 10.1126/science.153.3734.379 |
[11] | Lee CH , Wu SB , Hong CH , Yu HS , Wei YH . Molecular Mechanisms of UV-Induced Apoptosis and Its Effects on Skin Residential Cells: The Implication in UV-Based Phototherapy. Int J Mol Sci. (2013) ;14: (3):6414–35. doi: 10.3390/ijms14036414 |
[12] | Schuler M , Bossy-Wetzel E , Goldstein JC , Fitzgerald P , Green DR . p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. Journal of Biological Chemistry. (2000) ;275: (10):7337–42. doi: |
[13] | Kennedy SG , Kandel ES , Cross TK , Hay N . Akt/Protein kinase B inhibits cell death by preventing the release of cytochrome c from mitochondria. Molecular and cellular biology. (1999) ;19: (8):5800–10. doi: |
[14] | Cai Y , Luo Q , Sun M , Corke H . Antioxidant activity and phenolic compounds of 112 traditional Chinese medicinal plants associated with anticancer. Life sciences. (2004) ;74: (17):2157–84. doi: 10.1016/j.lfs.2003.09.047 |
[15] | Narendhirakannan R , Hannah CMA . Oxidative stress and skin cancer: an overview. Indian Journal of Clinical Biochemistry. (2013) ;28: (2):110–5. doi: 10.1007/s12291-012-0278-8 |
[16] | Gasparrini M , Forbes-Hernandez TY , Afrin S , Reboredo-Rodriguez P , Cianciosi D , Mezzetti B , et al Strawberry-based cosmetic formulations protect human dermal fibroblasts against UVA-induced damage. Nutrients. (2017) ;9: (6):605. doi: 10.3390/nu9060605 |
[17] | Parzonko A , Kiss AK . Caffeic acid derivatives isolated from Galinsoga parviflora herb protected human dermal fibroblasts from UVA-radiation. Phytomedicine. (2019) ;57: :215–22. doi: 10.1016/j.phymed.2018.12.022 |
[18] | Georgiev V , Ananga A , Tsolova V . Recent advances and uses of grape flavonoids as nutraceuticals. Nutrients. (2014) ;6: (1):391–415. doi: 10.3390/nu6010391 |
[19] | Bagchi D , Bagchi M , Stohs SJ , Das DK , Ray SD , Kuszynski CA , et al Free radicals and grape seed proanthocyanidin extract: importance in human health and disease prevention. Toxicology. (2000) ;148: (2-3):187–97. doi: 10.1016/S0300-483X(00)00210-9 |
[20] | Rasines-Perea Z , Teissedre P-L . Grape Polyphenols’ effects in human cardiovascular diseases and diabetes. Molecules. (2017) ;22: (1):68. doi: 10.3390/molecules22010068 |
[21] | Porcelli L , Iacobazzi RM , Quatrale AE , Bergamini C , Denora N , Crupi P , et al Grape seed extracts modify the outcome of oxaliplatin in colon cancer cells by interfering with cellular mechanisms of drug cytotoxicity. Oncotarget. (2017) ;8: (31):50845. doi: |
[22] | Agarwal C , Singh RP , Agarwal R . Grape seed extract induces apoptotic death of human prostate carcinoma DU145 cells via caspases activation accompanied by dissipation of mitochondrial membrane potential and cytochrome c release. Carcinogenesis. (2002) ;23: (11):1869–76. doi: 10.1093/carcin/23.11.1869 |
[23] | Reddivari L , Charepalli V , Radhakrishnan S , Vadde R , Elias RJ , Lambert JD , et al Grape compounds suppress colon cancer stem cells in vitro and in a rodent model of colon carcinogenesis. BMC complementary and alternative medicine. (2016) ;16: (1):278. doi: 10.1186/s12906-016-1254-2 |
[24] | Shirai A , Onitsuka M , Maseda H , Omasa T . Effect of polyphenols on reactive oxygen species production and cell growth of human dermal fibroblasts after irradiation with ultraviolet-A light. Biocontrol Sci. (2015) ;20: (1):27–33. doi: 10.4265/bio.20.27 |
[25] | Ndiaye M , Philippe C , Mukhtar H , Ahmad N . The grape antioxidant resveratrol for skin disorders: promise, prospects, and challenges. Archives of biochemistry and biophysics. (2011) ;508: (2):164–70. doi: 10.1016/j.abb.2010.12.030 |
[26] | Jang M , Cai L , Udeani GO , Slowing KV , Thomas CF , Beecher CW , et al Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science (New York, N.Y.). (1997) ;275: (5297):218–20. doi: 10.1126/science.275.5297.218 |
[27] | Hudson TS , Hartle DK , Hursting SD , Nunez NP , Wang TT , Young HA , et al Inhibition of prostate cancer growth by muscadine grape skin extract and resveratrol through distinct mechanisms. Cancer research. (2007) ;67: (17):8396–405. doi: 10.1158/0008-5472.can-06-4069 |
[28] | Engelbrecht AM , Mattheyse M , Ellis B , Loos B , Thomas M , Smith R , et al Proanthocyanidin from grape seeds inactivates the PI3-kinase/PKB pathway and induces apoptosis in a colon cancer cell line. Cancer letters. (2007) ;258: (1):144–53. doi: 10.1016/j.canlet.2007.08.020 |
[29] | Masuma R , Kashima S , Kurasaki M , Okuno T . Effects of UV wavelength on cell damages caused by UV irradiation in PC12 cells. Journal of photochemistry and photobiology. B, Biology. (2013) ;125: :202–8. doi: 10.1016/j.jphotobiol.2013.06.003 |
[30] | de Lima-Bessa KM , Armelini MG , Chigancas V , Jacysyn JF , Amarante-Mendes GP , Sarasin A , et al CPDs and 6-4PPs play different roles in UV-induced cell death in normal and NER-deficient human cells. DNA repair. (2008) ;7: (2):303–12. doi: 10.1016/j.dnarep.2007.11.003 |
[31] | Fiers W , Beyaert R , Declercq W , Vandenabeele P . More than one way to die: apoptosis, necrosis and reactive oxygen damage. Oncogene. (1999) ;18: (54):7719–30. doi: 10.1038/sj.onc.1203249 |
[32] | Rastogi RP , Richa , Sinha RP , Singh SP , Hader DP . Photoprotective compounds from marine organisms. Journal of industrial microbiology & biotechnology. (2010) ;37: (6):537–58. doi: 10.1007/s10295-010-0718-5 |
[33] | Besaratinia A , Yoon JI , Schroeder C , Bradforth SE , Cockburn M , Pfeifer GP . Wavelength dependence of ultraviolet radiation-induced DNA damage as determined by laser irradiation suggests that cyclobutane pyrimidine dimers are the principal DNA lesions produced by terrestrial sunlight. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. (2011) ;25: (9):3079–91. doi: 10.1096/fj.11-187336 |
[34] | Riou L , Eveno E , van Hoffen A , van Zeeland AA , Sarasin A , Mullenders LH . Differential repair of the two major UV-induced photolesions in trichothiodystrophy fibroblasts. Cancer research. (2004) ;64: (3):889–94. doi: 10.1158/0008-5472 |
[35] | Kris-Etherton PM , Hecker KD , Bonanome A , Coval SM , Binkoski AE , Hilpert KF , et al Bioactive compounds in foods: their role in the prevention of cardiovascular disease and cancer. The American journal of medicine. (2002) ;113: (Suppl 9B):71s–88s. doi: 10.1016/s0002-9343(01)00995-0 |
[36] | Jara-Palacios MJ , Hernanz D , Cifuentes-Gomez T , Escudero-Gilete ML , Heredia FJ , Spencer JP . Assessment of white grape pomace from winemaking as source of bioactive compounds, and its antiproliferative activity. Food chemistry. (2015) ;183: :78–82. doi: 10.1016/j.foodchem.2015.03.022 |
[37] | Barcia MT , Pertuzatti PB , Gomez-Alonso S , Godoy HT , Hermosin-Gutierrez I . Phenolic composition of grape and winemaking by-products of Brazilian hybrid cultivars BRS Violeta and BRS Lorena. Food chemistry. (2014) ;159: :95–105. doi: 10.1016/j.foodchem.2014.02.163 |
[38] | Niu S , Hao F , Mo H , Jiang J , Wang H , Liu C , et al Phenol profiles and antioxidant properties of white skinned grapes and their coloured genotypes during growth. Biotechnology & Biotechnological Equipment. (2017) ;31: (1):58–67. doi: |
[39] | Romano G , Briguori C , Quintavalle C , Zanca C , Rivera NV , Colombo A , et al Contrast agents and renal cell apoptosis. Eur Heart J. (2008) ;29: (20):2569–76. doi: 10.1093/eurheartj/ehn197 |




