Roles of Non-Coding RNAs as Novel Diagnostic Biomarkers in Parkinson’s Disease
Abstract
Parkinson’s disease (PD) is the second most common neurodegenerative disorder, affecting 5%of the elderly population. Currently, the diagnosis of PD is mainly based on clinical features and no definitive diagnostic biomarkers have been identified. The discovery of biomarkers at the earliest stages of PD is of extreme interest. This review focuses on the current findings in the field of circulating non-coding RNAs in PD. We briefly describe the more established circulating biomarkers in PD and provide a more thorough review of non-coding RNAs, in particular microRNAs, long non-coding RNAs and circular RNAs, differentially expressed in PD, highlighting their potential for being considered as biomarkers for diagnosis. Together, these studies hold promise for the use of peripheral biomarkers for the diagnosis of PD.
INTRODUCTION
Parkinson’s disease (PD) is the most common progressive neurodegenerative disorder, resulting from a pathophysiologic loss or degeneration of dopaminergic neurons in the substantia nigra of the midbrain and the development of neuronal Lewy bodies. Idiopathic PD is associated with risk factors including aging, family history, pesticide exposure, and environmental chemicals [1, 2]. Characterized by both motor and non-motor symptoms, PD patients classically display rest tremor, rigidity, bradykinesia, and stooping posture. PD can also be associated with neurobehavioral disorders (depression, anxiety) and cognitive impairment, that may emerge years prior to clinical diagnosis [3, 4]. Loss of dopaminergic neurons in the substantia nigra together with the presence of aggregates intracellular of protein deposits (named Lewy bodies) which are primarily composed of precipitates of the alpha-synuclein (α-syn) protein, are the neuropathological hallmark of sporadic PD. Although the causes remain unclear, sporadic PD likely results from a complex interaction of environmental/acquired and genetic/inherited factors [5]. Hereditary forms of PD represent only 5–10%of all cases, studies on these have provided indications on the pathogenesis, indeed, much of the knowledge of the genes and pathways involved in PD derives from studies on the hereditary forms of disease. To date, various PD-associated mutations have been identified in several genes. Out of the six genes undoubtedly linked to heritable, monogenic PD, mutations in SNCA and LRRK2 are responsible for autosomal-dominant PD forms, and mutations in PARKIN, PINK1, and DJ-1, are accountable for PD that displays an autosomal recessive mode of inheritance. Of note, SNCA is thought to be a crucial player in the pathogenesis of PD based on genetic, pathological, and cellular/molecular lines of evidence. PD diagnosis is still based on clinical criteria by identification of at least two neuromotor symptoms such as resting tremor, bradykinesia, rigidity and/or postural instability [6]. Another obstacle that presents with the diagnosis of PD is its phenotypic similarity to other atypical parkinsonian disorders. These include disorders such as progressive supranuclear palsy, multisystem atrophy, and corticobasal degeneration. These diseases affect the diagnosis of PD as the symptoms are overlapping and heterogeneous, especially in the early stages of the disease, making differentiation difficult. Therefore, obtaining an accurate diagnosis of PD is difficult during the early stages of the disease, and there is a high risk of misdiagnosis. Neuroimaging techniques, such as magnetic resonance imaging, [18F]fluorodopa positron emission tomography, single-photon emission computed tomography, and transcranial ultrasound have provided some utility in the diagnosis and staging of idiopathic PD but remain expensive and labor intensive. Blood and cerebrospinal fluid (CSF) have been extensively analyzed for protein biomarkers, dopamine metabolites, and amino acids. α-Synuclein comprises the main protein component of hallmark Lewy bodies and is measurable in blood CSF [7–9]. Despite advances in neuroimaging and genetics, the diagnosis of PD remains primarily clinical, and so, in recent years, the search for biomarkers to support early diagnosis has gained interest [8]. A biomarker is defined, by the National Institutes of Health Biomarkers Definitions Working Group, as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes or pharmacologic responses to a therapeutic intervention” [10]. Thus, biomarkers are useful for confirming diagnosis, prognosis, disease progression, response to therapy, through a qualitative and/or quantitative analysis and should be non-invasive, reproducible and specific to the disease under consideration. In this context, an alternative avenue is the development of peripheral biomarkers utilizing biofluids, which reflect molecular changes in the brain, and thus provide support for early diagnosis.
It is well known that RNA plays the fundamental role of mediator in the flow of genetic information from DNA to the final protein product. In recent years, a number of further biological functions of RNA have been identified, not only as structural and catalytic molecules, but also and above all as regulators of gene expression in most cellular processes: from differentiation to development. However, over the past decade, advances in whole genome analysis have shown that up to 90%of the human genome is transcribed, while tens of thousands of RNA transcripts have similar properties to mRNAs but are not translated into proteins. Indeed, approximately only 2%of RNA transcripts encode proteins, whereas the remaining transcripts are non-coding RNAs (ncRNAs). ncRNAs transcripts are further divided into housekeeping ncRNAs and regulatory ncRNAs. Housekeeping ncRNAs, which are usually constitutively expressed, include ribosomal, transfer, small nuclear, and small nucleolar RNAs. Regulatory ncRNAs are generally divided into two classes based on nucleotide length. Those less than 200 nucleotides are usually referred to as short/small ncRNAs and include microRNAs (miRNAs), small interfering RNAs, and PIWI-associated RNAs, whereas those greater than 200 nucleotides are known as long non-coding RNA (lncRNAs) [11]. Another class of regulatory ncRNAs are circular RNAs (circRNAs), molecules transcribed and spliced from exons in protein and noncoding genes [12]. ncRNAs representing a novel group of biomarkers and therapeutic targets in the pathophysiology of several neurodegenerative disorders. These expressed transcripts in the central nervous system (CNS) have been shown to regulate several signaling pathways implicated in the neurodegeneration, such as apoptosis and mitochondrial dysfunction [13, 14], and a number of lncRNAs, miRNAs and circRNAs also influence neurodegeneration in a direct manner [15, 16].
Due to their presence and relative stability in the bloodstream, often packed in extracellular vesicles like exosomes, ncRNAs appeared as potential disease markers [17], and promising therapeutic targets as they influence multiple biological pathways leading to the development and progression of disease. Several studies reported a permeability of the brain-blood barrier (BBB) in PD patients [18–20]. So, ncRNAs released from the brain into the blood may serve as biomarkers of pathology. In this study, we review the results of studies which evaluated the role of ncRNAs in the pathogenesis of PD.
NON-CODING RNAs FAMILY
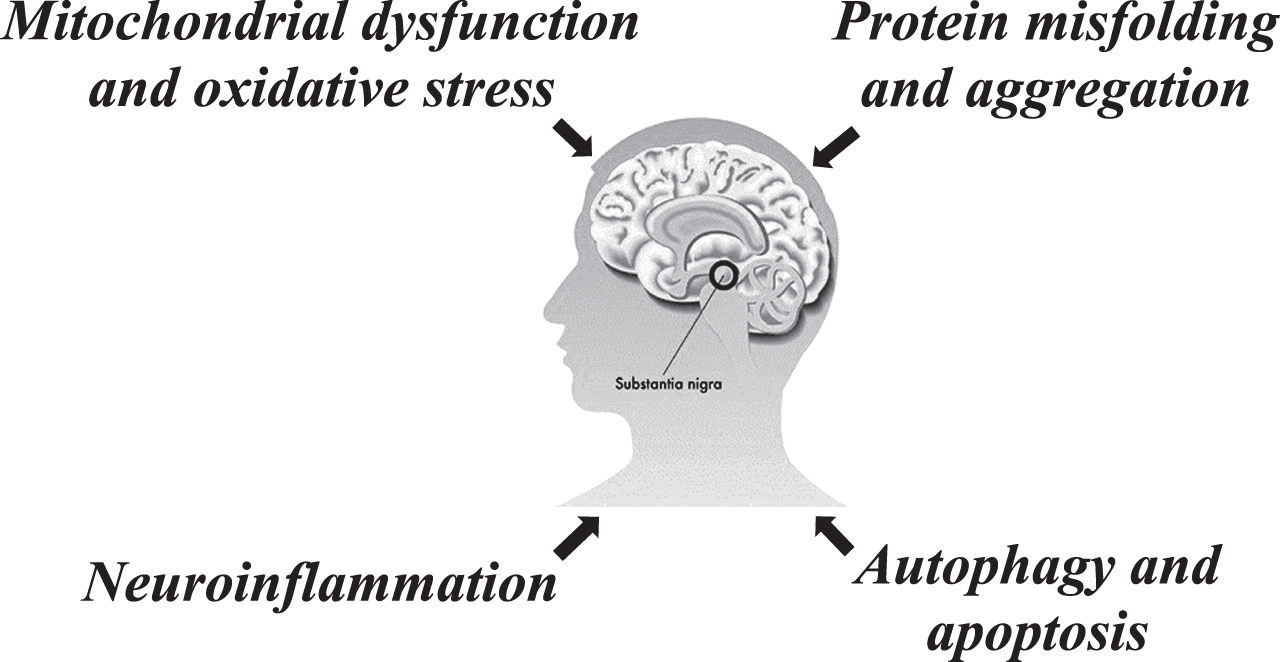
ncRNAs are a family of non-coding protein transcripts that modulate cell function by controlling gene expression programs through various mechanisms [21], and they includes lncRNAs (longer than 200 nucleotides), circRNAs (generated from pre-mRNA backsplicing), and miRNAs (around 21–25 nucleotides). Although most ncRNAs cannot be translated into protein, many function in the regulation of important biological processes by modulating transcription and post-translational modifications [22]. miRNAs, lncRNAs and circRNAs interact with each other, acting as competing endogenous RNAs (ceRNAs), and the latter, as is known, participate at the transcriptional and post-transcriptional level in the modulation of the target genes [23] linked to different pathway involved in PD (Fig. 1).
Fig. 1
Regulatory role of ncRNAs in Parkinson’s disease.
miRNAs
miRNAs are small ncRNAs that regulate gene expression by binding to the 3′-untranslated region of target RNAs, leading to the degradation or inhibition of translation, thereby affecting many regulatory processes (reviewed in [24]). The synthesis of these small RNA molecules occurs mainly through a canonical biogenesis pathway. Most of miRNAs genes are synthetized by RNA polymerase II into primary miRNA transcript and then cleaved into precursor miRNA (pre-miRNA) from which, after export to the cytoplasm, mature miRNA originates [25]. It is now clear the fundamental role of miRNAs in many biological functions. Indeed, it has been estimated that miRNAs may regulate the expression of about one-third of genes; moreover, single miRNA can regulate multiple transcripts, and using computational algorithms, several putative and targets for individual miRNAs were predicted [26]. In brain, miRNAs take part in spatiotemporal regulation of neuronal gene expression that is crucial for neural differentiation, circuit development, and modification of neuronal networks; therefore, an altered miRNAs expression can result in dysregulation of key genes and pathways that contribute to disease development [27]. An altered expression of miRNAs in the brain has been described in many neurodegenerative disorders [28], including PD. For the first time, Kim et al. studied the miRNAs expression profile in brain tissue of PD patients. Analyzing 230 miRNA precursors in the midbrain, cerebellum, and cortex samples, they observed a deficient expression of the precursor miR-133b in PD patient compared to healthy controls. They also associate a regulatory function of miR-133 on the maturation and function of dopaminergic neurons through a negative feedback circuit that includes the paired homeodomain transcription factor Pitx3 [29]. In a subsequent study, Schlaudraff and colleagues reanalyzed the expression of miR-133 in the entire midbrain tissue. They observed a reduction in miR-133 level in the midbrain of PD patients; on the contrary, no difference in the expression of miR-133 was found at the level of substantia nigra (SN) dopamine neurons (DA) between PD patients and controls [30]. Briggs et al. also investigate miRNAs expression in postmortem SN DA neurons in PD patients and controls. Using human microRNA TaqMan array the researchers identifying 159 dysregulated miRNAs (109 up- and 50 downregulated miRNAs) in PD patients with a different up and down trend between gender. Moreover, they found a network of miRNA target-genes linked to dysfunctional pathways related to PD [31]. Since, an altered miRNAs expression can result in dysregulation of key genes and pathways, the functional role of different miRNAs was investigated, and today several miRNAs are known to be implicated in PD-related pathways. For example, miR-7 inhibits α-Syn expression by binding to the 3′-UTR of its mRNA; furthermore, in differentiated ReNcell VM cells, it has been observed that miR-7 improve α-Syn and its aggregate clearance by promoting autophagy [32]. Also, miR-10a has been suggested as α-Syn regulator. A decreased level of miR-10a has been detected in the midbrain of A30P α-syn transgenic mice and in SH-SY5Y cells overexpressing A30P α-syn. Moreover, it was demonstrated that miR-10a attenuated α-syn aggregation and toxicity through suppression of proapoptotic protein BCL2L11 in SH-SY5Y cells [33]. The leucine-rich repeat kinase 2 (LRRK2) gene has been found to cause both familial and sporadic PD [34], although the molecular mechanisms underlying its action are not yet clear. By in silico analysis Cho and colleagues identified a miR-205 binding site in the 3’-UTR region of the LRRK2 gene. Through in vitro studies, they investigated the role of miR-205 in the regulation of LRRK2 expression, observing a dose-dependent reduction in LRRK2 protein level in primary neuron cultures. Furthermore, they found an inverse correlation between frontal cerebral cortex and striatum of patients with sporadic PD and controls [35]. DJ-1 is another target of miRNAs, as well asmiR-4639-5p may regulate the DJ-1 expression through binding the 3’-UTR of DJ-1 mRNA, resulting in downregulation of protein level with consequent oxidative stress and neuronal death [36]. The study of Wang et al. suggests that upregulation of miR-124 could regulate apoptosis and impaired autophagy process in the MPTP model of PD, thereby reducing the loss of DA neurons. Indeed, miR-124 could have a neuroprotective effect by targeting Bim a BH3-only protein and consequently preventing the translocation of Bax to the mitochondrion [37]. Although most miRNAs are brain-specific, they have been found circulating in different biofluids [38], both excreted within extracellular vesicles, such as exosome [39] and excreted as free miRNAs associated with diverse proteins such as Argonaute2 [40]. Due to their capacity to cross the BBB, miRNAs have the potential to be valuable biomarkers, providing information on the pathological state detected in the CNS [41]. Noteworthy disclosures regarding published circulating miRNAs in PD patients are found in numerous recent reviews. Certainly, blood and its derivatives are the main sources towards which the search for non-invasive miRNA biomarkers for PD has been directed, while only in a few works the expression of miRNAs in CSF, has been investigated [42–44], considering the invasiveness of lumbar puncture.
From studies it emerges that several dysregulated miRNAs have been detected in different biological fluids of patients with PD, but few of these miRNAs have been found significantly dysregulated in multiple studies, and in some cases even with opposite results (Table 1). Ultimately, there is not always overlap across the studies. The lack of overlap could be due to the fact that the variables in these studies are many, such as the nature of the biological liquid (e.g., CSF, whole blood, blood components), the methods for exosome isolation (e.g., ultracentrifugation, commercial kits), and the methods of miRNAs detection (e.g., qRT-PCR, microarrays, RNA sequencing). In this regard, thanks to its sensitivity and specificity, actually, qRT-PCR is the gold standard for miRNAs quantification, but it is also used for the validation of high-throughput profiling. Among the high-throughput technologies, microarrays provide comprehensive coverage, but limited accuracy, while NGS technologies allow the identification of novel miRNAs, but their main disadvantage is complex data interpretation and analysis as well as high costs [61]. For studies investigating the levels of miRNAs encapsulated in exosomes, another variable to consider is the method used to isolate the exosomes: ultracentrifugation is commonly used to isolate exosome, and the use of easy-to-use precipitation solutions or commercial kits as they do not require expensive equipment. However, it is known that difference in isolation methods can cause variations in the concentration, purity, and size both of the exosomes and exosomal RNA [62]. Therefore, a standardization of these variables would be useful in identifying reliable biomarkers.
Table 1
miRNAs involved Parkinson’s disease
| miRNAs | Source | Cohort composition | Methods | Regulation | Reference |
| miR-29c-3p | |||||
| PBMCs | 19 PD | microarrays | ↓ | [45] | |
| 13 HC | |||||
| serum | 10 PD | TLDA/qRT-PCR | ↓ | [46] | |
| 10 HC | |||||
| 20 PD | |||||
| 20 HC* | |||||
| 65 PD | |||||
| 65 HC** | |||||
| serum | 138 PD | qRT-PCR | ↓ | [47] | |
| 112 HC | |||||
| serum | 75 PD | qRT-PCR | ↓ | [48] | |
| 77 HC | |||||
| serum | 51 PD | qRT-PCR | ↑ | [49] | |
| 20 HC | |||||
| miR-19b-3p | |||||
| PBMCs | 19 PD | microarrays | ↓ | [45] | |
| 13 HC | |||||
| CSF | 57 PD | NGS | ↑ | [50] | |
| 65 HC | |||||
| serum | 10 PD | TLDA/qRT-PCR | ↓ | [46] | |
| 10 HC, | |||||
| 20 PD | |||||
| 20 HC* | |||||
| 65 PD | |||||
| 65 HC** | |||||
| CSF exosome | 47 PD | TLDA/qRT-PCR | ↓ | [51] | |
| 27 CTRL | |||||
| 78 PD | |||||
| 35 CTRL | |||||
| Serum exosome | 109 PD | qRT-PCR | ↓ | [52] | |
| 40 HC | |||||
| miR-29a-3p | |||||
| whole | 8 PD | qRT-PCR | ↓ | [53] | |
| blood | 8 HC | ||||
| serum | 10 PD | TLDA/qRT-PCR | ↓ | [46] | |
| 10 HC, | |||||
| 20 PD | |||||
| 20 HC* | |||||
| 65 PD | |||||
| 65 HC** | |||||
| PBMCs | 38 PD | qRT-PCR | ↑ | [54] | |
| 38 HC | |||||
| serum | 78 PD | qRT-PCR | ↓ | [48] | |
| 80 HC | |||||
| miR-24-3p | |||||
| Serum | 6 PD | TLDA/qRT-PCR | ↑ | [55] | |
| 5 HC | |||||
| 25 PD* | |||||
| 25 HC | |||||
| CSF | 28 PD | qRT-PCR | ↓ | [56] | |
| 28 PD | |||||
| Serum | 109 PD | qRT-PCR | ↑ | [52] | |
| exosome | 40 HC | ||||
| miR-335-5p | |||||
| PBMCs | 19 PD | microarrays | ↓ | [45] | |
| 13 HC | |||||
| Serum | 16 PD | microarrays | ↑ | [57] | |
| 8 HC | qRT-PCR | ||||
| Serum | 20 PD | qRT-PCR | ↓ | [58] | |
| 20 HC | |||||
| 20 PD | |||||
| 20 HC | |||||
| miR-433-3p | |||||
| CSF | 57 PD | NGS | ↓ | [50] | |
| 65 HC | |||||
| Plasma | 46 PD | qRT-PCR | ↓ | [59] | |
| 49 HC | |||||
| Plasma | 99 PD | qRT-PCR | ↑ | [60] | |
| 100 HC |
PBMC, peripheral blood mononuclear cells; CSF, cerebrospinal fluid; PD, Parkinson’s disease; HC, healthy controls; TLDA, TaqMan low-density array; qRT-PCR, quantitative Reverse Transcription PCR; NGS, Next Generation Sequencing; *first cohort of validation; **second cohort of validation; ↑ or ↓ symbolize up- or downregulation of miRNAs expression.
Actually, exosome-based biomarkers represent an emerging non-invasive source for researching potential miRNA as biomarkers of disease [63]. The exosomes are nanovesicles released from cells into the biofluids and involved in cell-cell communication. They deliver their cell- or condition-specific cargo of proteins, lipids, and genetic materials, including ncRNAs [64], and represent an enriched source of miRNAs protected by RNase [65]. Therefore, due to their features and ease of isolation exosomal miRNAs have been investigate as possible biomarkers for PD.
For the first time Gui et al. analyzed exosomal miRNAs expression in CSF of PD patients (n = 47) comparing to healthy controls (n = 27) and Alzheimer’s disease patients (n = 28). They found that miR-1 and miR-19b-3p were significantly downregulated, while miR-153, miR-409-3p, miR-10a-5p, and let-7g-3p where significantly upregulated also in independent cohort; moreover, the ROC curve analysis confirmed their capability to discriminate PD patients and healthy controls with good specificity and sensitivity [51]. Investigating the potential of exosomal miRNAs in plasma, Yao et al. identified two dysregulated miRNAs in PD patients, in particular miRNA-331-5p were significantly higher, and in reverse miRNA-505 was lower in PD compared to healthy controls, with an AUC 0.849 and 0.898, respectively [66].
Dos Santos et al. searched for exosomal miRNAs at the earliest stage of PD. Using Next Generation Sequencing (NGS) to analyze exosomal miRNAs in the CSF and then a machine learning approach, they identify a miRNAs panel (Let-7f-5p, miR-125a-5p, miR-151a-3p, miR-27a-3p, and miR-423-5p) that could discriminate early stage PD patients from controls with an AUC of 82%[67].
Although there are still not many studies in this field, to date these results seem encouraging, and suggest the continuation of further studies in this direction. Another important consideration regarding the potential of miRNAs as biomarkers for PD, concerns the size and ethnicity of the study cohorts. Several promising miRNAs have been identified, but the use of larger and independent validation cohorts could make them more robust classifiers of the disease. In particular, Patil et al. investigated a miRNAs profile across geographically diverse cohorts [57].
lncRNAs
lncRNAs are defined as a heterogeneous class of ncRNAs, canonically expressed under a linear form, longer than 200 nucleotides, and without protein coding capabilities. Their biogenesis is similar to that of mRNAs with RNA Polymerase II used for their transcription. The transcription process, however, is often subject to capping, canonical and alternative splicing, as well as polyadenylation. They are located in the nucleus or cytoplasm. In the nucleus, they can specifically induce gene silencing [68], and in the cytoplasm, they can serve as ceRNAs to modulate miRNAs expression [69]. They can also control transcriptional activity by directly or indirectly targeting mRNAs [70]. Based on the relative positions of the lncRNAs coding sequences and protein-coding genes, they are categorized as: a) sense lncRNAs overlapping with the protein-coding genes; b) antisense lncRNAs overlapping with the antisense strands of protein-coding genes; c) bidirectional lncRNAs transcribed from the divergent bidirectional promoters relative to the protein-coding genes; d) intronic lncRNAs derived entirely from the introns of transcripts; and e) intergenic lncRNAs sequences located between but not overlapping with the protein-coding genes. Studies have shown that lncRNAs are involved in transcriptional and epigenetic mechanisms, gene regulation in post-transcription level, alternative splicing, and in gene silencing (reviewed in [71]), although the number of well characterized lncRNAs, to date, is limited. Recently, lncRNAs have received widespread attention due to their diverse roles in brain development, neuronal function, maintenance and differentiation [72], and neurological diseases (reviewed in [73]). Existing studies have confirmed that lncRNAs are highly expressed in various parts of both the CNS and the brain [74], and the abnormal expression of lncRNAs is closely associated with PD, as described elsewhere [75]. To date few studies focused on brain tissues in PD, in particular an abnormal expression profiles of lncRNAs were found in the SN and in various parts of brain of PD patients. Kraus et al. found five lncRNAs that were differentially expressed in PD, in particular lncRNA-p21, MALAT1, SNHG1, and NEAT1 were significantly upregulated, whereas H19 upstream conserved 1 and 2 were significantly downregulated. Of note, these dysregulated lncRNAs appeared in the early stages of PD and preceded the course of the disease [76]. Finally, elevated levels of lncRNA NEAT1 have been described in the peripheral blood of PD patients [77]. Ni et al. identified significant changes in the expression of 87 lncRNA in the SN of patients with PD compared to normal tissues, among which the significantly upregulated lncRNA AL049437 likely contributed to the risk of PD, whereas the downregulated lncRNA AK021630 likely inhibited PD development [78]. Since the function of lncRNAs is related to their cellular localization as well as their expression, a genome-wide approach to determine the identification of lncRNAs could greatly improve our understanding of their role in pathophysiology of disease. The research on the role of lncRNAs in PD is limited at present, but several studies have shown the involvement of lncRNAs in the different pathophysiological mechanisms underlying PD, including alpha- synuclein aggregate, calcium homeostasis, axonal transport, oxidative stress, mitochondrial dysfunction, and neuroinflammation (reviewed in [79]). Furthermore, since lncRNAs can act both by activating and repressing gene expression, they could represent key components in the regulation of PD-linked genes. About that, in a recent study Elkouris et al. show that a number of lncRNAs may alter the expression of PD-linked genes, thus establishing, for the first time, a correlation between PD-affected brain regions and specific lncRNAs. Indeed, they mainly focused on genes linked to familial PD, such as UCH-L1, PINK1, DJ1, SNCA, and LRRK2, of note the contribution of the latter two genes to the pathogenesis of the disease is strongly supported by genetic studies. In addition, the authors also chose the GBA1 and MAPT genes, considered susceptibility genes. Interestingly, six lncRNAs (SNCA-AS1, AK127687, UCHL1-AS1, PINK1-AS1, AX747125, and MAPT-AS1 are found under expressed in the SN, and of these 6, three (AK127687, UCHL1-AS1, and MAPT-AS1) are found under expressed in the cerebellum of PD patients compared to controls [80]. In support of these findings, previous studies have reported that MAPT-AS1, PINK1-AS1, and UCHL1-AS1 affect expression levels of MAPT, PINK1, and UCHL1 genes, respectively [81, 82]. Of note, MAPT-AS1 was also recently reported to be under-expressed in many brain regions of PD affected patients, including putamen, anterior cingulate cortex, visual cortex, and cerebellum [83]. Recently, to emphasize the importance of lncRNAs in PD, results of RNA sequencing and microarray screening have shown that numerous lncRNAs are differentially expressed in the peripheral blood of patients with PD. Specifically, five studies used peripheral blood samples to screen lncRNA expression profiles between PD patients and normal controls. Soreq et al. employed whole-transcriptome RNA sequencing to screen leukocyte-expressed lncRNAs in patients with PD and controls and found 13 lncRNAs which differential expressed. They found that the expression of five lncRNAs was upregulated in the disease, including the spliceosome component U1, supporting the idea that splicing modulations are involved in PD pathogenesis, and RP11-462G22.1 (lnc-FRG1-3), related to the muscle rigidity that occurs in PD. Elsewhere, the differential expression of RP11-462G22.1 was confirmed when comparing CSF exosomes from PD patients and healthy controls [51, 84]. Zhou et al. performed RNA sequencing and bioinformatics analysis to obtain differentially expressed mRNAs and lncRNAs between patients with PD and normal controls, and they identified a total of 857 differentially expressed mRNAs (304 upregulated and 553 downregulated) and 77 differentially expressed lncRNAs (38 upregulated and 39 downregulated). The authors point out that gene expression results validated by datasets GSE57475 and GSE68719, downloaded from the Gene Expression Omnibus (GEO), were consistent with their RNA-sequencing results [85]. Chi et al. performed microarray analysis of dataset GSE6613, downloaded from the GEO, containing blood samples of 22 healthy controls and 50 patients with PD, and they found seven differentially expressed lncRNAs, in particular two upregulated (LINC00302 and LINC00328) and five downregulated (FAM215A, MCF2L-AS1, NOP14-AS1, PART1, and XIST) lncRNAs. Among them, XIST, the most vital regulator of X chromosome in mammals, was markedly decreased in PD, it is known that lncRNA-XIST regulate the cell proliferation and apoptosis by modulating the MAPK pathway associated with the PD pathogenesis, albeit further research is warranted to clarify its mechanism of action in PD [86]. Wang et al. studied the differential expression of lncRNAs in peripheral blood exosomes, all lncRNAs obtained from exosomal lncRNAs from PD patients and healthy control subjects were deep sequenced via next generation sequencing and real-time quantitative polymerase chain reaction, and they found 15 upregulated and 24 downregulated exosomal lncRNAs in the PD group. Furthermore, according to the lncRNA
differential expression results, the authors found MSTRG.336210.1 and lnc-MKRN2-42:1 were highly expressed among healthy subjects, while MSTRG.242001.1 and MSTRG.169261.1 were highly expressed among PD patients. Finally, they selected lnc-MKRN2-42:1 for further correlation analysis between this lncRNA and clinical characteristics of PD patients, finding it a positively correlated with the Movement Disorder Society-Sponsored Revision Unified Parkinson’s Disease Rating Scale (MDS-UPDRS) III score for patients with PD and, thus, involved in the onset and development of the disease [87]. Fan et al. profiled lncRNA and mRNA expression in circulating leukocytes using microarray analysis. They identified 122 differentially expressed lncRNAs, including 95 upregulated and 27 downregulated lncRNAs, and 48 mRNAs between the circulating leukocytes from PD patients and healthy controls. Gene function and pathway analysis of the 48 deregulated mRNAs revealed biological pathways related to PD pathogenesis, including immune response, inflammatory response, MAPK, and Jak-STAT pathway. To underline that, the upregulation of four lncRNAs (AC131056.3-001, HOTAIRM1, lnc-MOK-6:1, and RF01976.1-201) in circulating leukocytes of PD patients were further confirmed in a larger cohort of patients. Furthermore, they demonstrated that the overexpression of lncRNA AC131056.3-001 or HOTAIRM1 could reduce cell viability and promote apoptosis of DA neurons in SHSY5Y cells [88]. All these results come from preliminary studies of high-throughput RNA sequencing and microarray, have been confirmed, albeit in small cohorts of patients, by RT-qPCR, the most sensitive method to accurately determine expression changes between cohorts. Taken together all these results demonstrate that there were numerous lncRNA differentially expressed in brain tissues and peripheral blood of PD patients. In order to understand fully the value of these findings, future studies should address in larger cohorts of patients, independent and of different ethnicity. A summary of the expression of longRNAs in brain and blood samples of PD patients are presented in Table 2. Finally, several studies have shown how lncRNAs play an important role both in the biological mechanisms underlying PD and in the regulation of multiple molecular pathways, such as protein misfolding and aggregation, mitochondrial disfunction, oxidative stress, neuroinflammation, autophagy, and apoptosis (Fig. 1). A nonlysosomal pathway of protein degradation is the ubiquitin-proteasome system (UPS), which removes damaged and aberrant proteins in the cells, plays an important role in the pathogenesis of PD. It is known that intraneuronal α-synuclein protein aggregates, observed in all patients with PD, are degraded by the action of the UPS and the lysosomal autophagy system [89]. Ubiquitin carboxy-terminal hydrolase L1 (Uchl1) is a gene involved in the UPS of PD, and the lncRNA UCHL1-AS1, by increasing synthesis of the UCHL1 protein, directly affects the translation of the UCHL1 protein, leading to disruption of the UPS [90]. It was demonstrated that mitochondrial dysfunction is a key element in PD pathogenesis, furthermore, mitochondrial dysfunction leads to oxidative stress, which is observed at a high level in the brain of PD patients [91]. Recently, it has been shown that the lncRNA NEAT1, which is overexpressed in the substantia nigra of PD, plays a neuroprotective role against drug-induced oxidative stress [92]. Autophagy and apoptosis play an important role in the PD processes, in particular apoptosis is considered to be an essential signal for dopaminergic neuronal degradation in PD, and it is closely related to mitochondrial dysfunction and oxidative stress. lncRNA MALAT1 regulates the MPP+-induced apoptosis of the MN9D cells by inhibiting miR-205-5p targeting LRRK2 [93]. Another important factor of PD is the neuroinflammation: the inflammatory cytokines, secreted by the glial cells, participate in neuroinflammatory responses to induce apoptosis of the dopaminergic neurons. In human blood, microarray technology used to detect the differentially expressed genes and lncRNAs associated with PD showed that genes, downregulated, differentially expressed in the regulatory network were enriched in the immune response [86].
Table 2
lncRNAs in brain and blood samples of PD patients
| lncRNA | Regulation | Cohort composition | Tissue/model | Methods | Reference |
| lincRNA-p21 | ↑ | 20 PD vs 10 controls | Human brain specimens | Real-time PCR validation | [74] |
| MALAT1 | ↑ | ||||
| SNHG1 | ↑ | ||||
| NEAT1 * | ↑ | ||||
| H19 | ↓ | ||||
| MAPT-AS1 | ↓ | 10 PD vs 10 controls | Tissue samples | Real-time PCR validation | [81] |
| SNCA-AS1 | ↓ | 9 PD vs 8 controls | Tissue samples | Real-time PCR validation | [78] |
| AK127687 | |||||
| UCHL1-AS1 | |||||
| PINK1-AS1 | |||||
| AX747125 | |||||
| MAPT-AS1 | |||||
| AL049437 | ↑ | 11 PD vs 14 controls | Tissue samples | Real-time PCR validation | [76] |
| AK021630 | ↓ | ||||
| U1 | ↓ | 3 PD vs 3 controls | Blood leukocytes | Real-time PCR | [82] |
| RP11-462G22.1 | ↓ | ||||
| RP11-462G22.1 | ↑ | 47 PD vs 27 controls | Cerebrospinal fluid | Real-time PCR validation | [51] |
| AC131056.3-001 | ↑ | 72 PD vs 22 controls | Blood leukocytes | Real-time PCR validation | [86] |
| HOTAIRM1 | ↑ | ||||
| lnc-MOK-6:1 | ↑ | ||||
| RF01976.1-201 | ↑ | ||||
| NEAT1* | ↑ | 61 PD vs 42 controls | Peripheral blood mononuclear cells | Real-time PCR validation | [75] |
| linc00302 | ↑ | 50 PD vs 22 controls | Blood leukocytes | Real-time PCR validation | [84] |
| linc00328 | ↑ | ||||
| FAM215A | ↓ | ||||
| MCF2L-AS1 | ↓ | ||||
| NOP14-AS1 | ↓ | ||||
| PART1 | ↓ | ||||
| XIST | ↓ | ||||
| MSTRG.336210.1 | ↑ | 13 controls | Blood exosomes | Real-time PCR validation | [85] |
| lnc-MKRN2-42:1 | ↑ | ||||
| MSTRG.242001.1 | ↑ | ||||
| MSTRG.169261.1 | ↑ | 32 PD |
PD, Parkinson’s disease; *lncRNAs found to be differentially expressed in more than one cohort; ↑ or ↓ symbolize up- or downregulation of lncRNAs expression.
circRNAs
Recently, cirRNAs, another type of ncRNA with covalently closed ends has gained attention. circRNAs are known to form through a back-splice reaction, which mainly consists of three mechanisms: 1) intron coupling-driven circularization, where RNA circularization can be generated through direct base coupling between flanking introns; 2) RBP-driven circularization, where the back-splicing event can be guided by the RNA-binding proteins (RBPs) to recognize and cling to the specific motif of the introns flanking the circularized exons; 3) lariat driven circularization, which occurs in the process of linear RNA splicing and through which, circRNAs are formed either from intron removal or from exon-skipping events [94]. There are different subtypes of circRNAs, including exonic, intronic, and exo-intronic. Exonic circRNAs are mostly localized in the cytoplasm, where they act as sponges for miRNAs, and RBPs, thus inhibiting their interaction with mRNA targets [95], on the contrary, intronic or exo-intronic circRNAs are mostly located in the nucleus and have few or no binding site for miRNAs, but act on transcriptional control [96]. The result is a non-polyadenylated circular transcript, and the lack of free ends, which are normally targeted by 3’ and 5’ exoribonucleases, makes circRNAs, compared with linear ncRNAs, more resistant to RNA exonuclease and exceptionally stable in cells [97]. circRNAs, a new class of single-stranded regulatory RNAs, are involved in different mechanisms of action in both physiological and pathological conditions, and recent studies revealed that many circRNAs play important roles in the regulation of gene expression at both transcriptional and post-transcriptional levels, however, the functions are not yet fully understood [98, 99]. circRNAs have been shown to be largely conserved and more abundant in the brain and exosomes than in other tissues [100, 101], consequently, circRNAs play an important role in the pathogenesis and progression of neurodegenerative diseases [102]. Due to their characteristics of crossing the BBB, being more resistant, than linear RNA, to RNA endonuclease and stable in cells, they are good candidates as potential diagnostic markers for CNS disorders. Finally, being also present in the human blood, they provide information about the disease status in the CNS. Many brain-enriched circRNAs have been associated with pathogenetic processes of neurodegeneration, as ciRS-7, highly abundant in the brain, is down-regulated in the brain of patients with Alzheimer’s disease, and acts as a sort of endogenous, competitive and anti-complementary miRNA “sponge” to adsorb, and therefore block, the normal functions of miRNA-7 [103]. More recently, Hanan et al. found that another circRNA, circSLC8A1, is increased in the substantia nigra of patients with PD and in cultured cells exposed to the oxidative stress-inducing agent; on the contrary were decreased in cells treated with the neuroprotective antioxidant [104]. Importantly, circSLC8A1 carries seven binding sites for miR-128, an abundant and brain-restricted miRNA that governs neuronal excitability and motor behavior [105]. In addition, another circRNA, called circSNCA, can sponge miR-7, thereby upregulating expression of SNCA mRNA, resulting in reduced autophagy and increased apoptosis in SH-SY5Y cells [106]. An advantageous strategy for understanding neurodegenerative diseases such as PD is the comprehensive study of the functions of circRNAs in different regions of the brain. About this, Jia et al. performed RNA sequencing of varying brain regions to construct circRNAs expression profiles of the cerebral cortex (CC), hippocampus (HP), striatum (ST), and cerebellum (CB) of PD mouse model. The results of this study showed circRNAs to be expressed differently in the CC, HP, ST, and CB, and found many specific circRNAs in each region, of which mmu_circRNA_0001320 was highly expressed in CB, while mmu_circRNA_ 0004144, mmu_circRNA_0000468, and mmu_circRNA_0013321 were not highly expressed in ST [107]. Indeed, this suggest that circRNAs, being tissue-specific, play important physiological and pathological roles and may serve as potential diagnostic biomarkers or as therapeutic targets for disease [108]. circRNAs, due to their covalently closed-loop structures, can resist degradation by RNases, making them highly stable in plasma, serum or other biofluids, and a number of highly abundant circRNAs have been found to exist in human peripheral blood. Memczak and colleagues were among the first to describe the presence of circRNAs in the blood [109] and other groups detecting circRNAs in other body fluids, as well as in exosome [101, 110]. Peripheral cells and in particular peripheral blood mononuclear cells (PBMCs) appear to participate directly in neurodegenerative processes as they inherit the same genetic information as brain cells. PBMCs thus represent a window on the CNS and are of particular importance for all those neurodegenerative diseases in which the affected tissue is not directly accessible for study [111–113]. Thus, PBMCs represent a powerful and non-invasive tool for the identification of new diagnostic biomarkers. In this regard, Ravanidis et al. profiled brain-enriched circRNAs in peripheral blood from control subjects and patients with PD using a RT-qPCR-based. They considered 87 brain-enriched circRNAs from which more than half were confidently detected in PBMCs and, in particular, found six circRNAs differentially expressed in PD with a 17%decrease on average from healthy control subjects levels [114]. Although more research is still needed to confirm these findings, dysregulated circRNAs form a robust set of brain-associated circRNA which may be further evaluated as diagnostic and possible therapeutic targets for PD. Regarding clinical application, it is important that the study population is phenotypically well defined. Last, but not least, to emphasize their biological role, in silico analysis can provide a comprehensive guide to the pathways and processes they control [114].
CONCLUDING REMARKS
PD is a neurodegenerative disease characterized by a movement disorder and the main neuropathological feature includes a loss of nigrostriatal dopaminergic neurons. PD diagnosis mainly based on the history of the disease and clinical manifestations. At present, the understanding of the underlying pathogenesis of the disease is not entirely clear, and although drugs reduce symptoms, they are unable to prevent disease progression. Consequently, it is critical for the choice of targeted therapy to study the molecular mechanisms underlying PD. Early disease biomarkers are still unknown and new future genetic targets are urgently needed. It is known that about 2%of the human genome is coding, while the remainder represents the non-coding portion of RNA, which however is thought to have a critical regulatory activity in the normal cell development, function, and pathogenesis of various diseases, in particular neurodegenerative disorders like PD [115]. These ncRNAs are classified as small and long non-coding. The most studied classes of small ncRNAs are miRNAs. Several works have highlighted the presence of miRNA in different biological fluids of PD patients, albeit with different expression profiles. In this regard, it should be emphasized that many factors influence the expression of circulating miRNAs, such as the choice of the type of body fluid, major study cohorts, even the choice of methodologies such as qRT-PCR or NGS sequencing platforms. However, precisely because of their stability characteristics in various biological fluids and the fact that they can be accurately quantified by routine and fast laboratory methods, they could represent promising diagnostic biomarkers for PD. Another class of non-coding RNA involved in transcriptional and epigenetic mechanisms are lncRNAs. Although research on lncRNAs is at an early stage, scientific data show that some lncRNAs are differently altered over time in the brains of patients with PD [76]. Recent studies have used RNA sequencing data analysis to evaluate lncRNA levels differentially expressed in leukocytes of patients with PD compared to controls, as well as in the human SN of PD patients. These studies highlighting the importance of lncRNAs as diagnostic tools [116]. Indeed, miRNAs/lncRNAs play an important role in the pathogenesis of PD. Thanks to the continuous publication of research results, greater attention is given to the possibility of selecting specific miRNA/lncRNA as biomarkers for the clinical diagnosis of PD. However, it is necessary to investigate and elucidate the regulatory mechanisms of miRNA/lncRNA in PD in order to improve the applicability and accuracy of these molecules as biomarkers for clinical diagnosis. Recent studies have focused on a new class of non-coding RNA expressed as a single-stranded circular transcript, covalently closed, due to the lack of free ends, which are usually attacked by ribonucleases, and it is precisely this peculiarity that makes them extremely stable. These circRNAs are now recognized as having important biological roles; they are widely conserved and more abundant in the brain than in other tissue. To date, few studies have evaluated the role of circRNAs in PD suggesting that they may interfere with downstream targets. For example, ciRS-7 it has been identified as a sponge for miR-7, of note, high neuronal α-synuclein expression is implicated in PD, and SNCA is a target gene of miR-7. Another circRNA, circSLC8A1, was found to increase in the substantia nigra of PD patients, and this circRNA carrier sites for miR-128, an abundant miRNA that regulates neuronal excitability. Taken together, these studies provide insights into the possible functional role of circRNAs in PD development. The availability of circRNAs in body fluids identifies them as potential disease biomarkers. As is known, BBB is compromised in neurodegenerative diseases, such as PD [117], and this would allow circRNAs encapsulated in extracellular vesicles, exosomes, or free to reach the peripheral circulation. This situation leads us to correlate the blood levels of specific neuronal circRNAs with the disease, with its progression and possibly with the response to treatments. In summary, based on the results obtained in these studies, ncRNAs have the potential to become useful biomarkers, and can help unravel the complex pathophysiological mechanisms that underlie PD. However, many factors contribute or influence the expression levels of these molecules, and these factors need to be considered. The type of body fluid used for the study is important as inconsistencies in expression levels are observed when comparing CSF, free blood cells, and blood cells. Also, methodological and technical implications such as the sequencing platform (e.g., NGS vs. qRT-PCR), isolation and purification of the samples and data normalization need to be considered. Additionally, there are other factors such as ethnicity, age, and gender. Finally, the need for larger study cohorts and dedicated studies to validate these findings in cohorts with different neurodegenerative diseases. In conclusion, the synergy between these novel experimental approaches combined with other biomarkers and imaging tools will be of great importance to define the role of these ncRNAs as novel biomarkers in PD.
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
REFERENCES
[1] | Kalia LV , Lang AE ((2015) ) Parkinson’s disease. Lancet 386: , 896–912. |
[2] | Lew M ((2007) ) Overview of Parkinson’s disease. Pharmacotherapy 27: , 155–160. |
[3] | Mazzoni P , Shabbott B , Cortés JC ((2012) ) Motor control abnormalities in Parkinson’s disease. Cold Spring Harb Perspect Med 2: , a009282. |
[4] | Teive HA , Bertucci Filho DC , Munhoz RP ((2016) ) Unusual motor and non-motor symptoms and signs in the early stage of Parkinson’s disease. Arq Neuropsiquiatr 74: , 781–784. |
[5] | Shulman JM , De Jager PL , Feany MB ((2011) ) Parkinson’s disease: Genetics and pathogenesis. Annu Rev Pathol 6: , 193–222. |
[6] | Balestrino R , Schapira AHV ((2020) ) Parkinson disease. Parkinsons Dis 27: , 27–42. |
[7] | Marques O , Outeiro TF ((2012) ) Alpha-synuclein: From secretion to dysfunction and death. Cell Death Dis 3: , 350. |
[8] | Miller DB , O’Callaghan JP ((2015) ) Biomarkers of Parkinson’s disease: Present and future. Metabolism 64: , S40–S46. |
[9] | Emamzadeh FN , Surguchov A ((2018) ) Parkinson’s disease: Biomarkers, treatment, and risk factors. Front Neurosci 12: , 612. |
[10] | Biomarkers Definitions Working Group ((2001) ) Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin Pharmacol Ther 69: , 89–95. |
[11] | Maxmen A ((2013) ) RNA: The genome’s rising stars. Nature 496: , 127–129. |
[12] | Yang Y , Fan X , Mao M , Song X , Wu P , Zhang Y , Jin Y , Yang Y , Chen LL , Wang Y , Wong CC , Xiao X , Wang Z ((2017) ) Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 27: , 626–641. |
[13] | Niu M , Xu R , Wang J , Hou B , Xie A ((2016) ) MiR-133b ameliorates axon degeneration induced by MPP(+) via targeting RhoA. Neuroscience 325: , 39–49. |
[14] | Kim J , Fiesel FC , Belmonte KC , Hudec R , Wang WX , Kim C , Nelson PT , Springer W , Kim J ((2016) ) miR-27a and miR-27b regulate autophagic clearance of damaged mitochondria by targeting PTEN-induced putative kinase 1 (PINK1). Mol Neurodegener 11: , 55. |
[15] | Wu YY , Kuo HC ((2020) ) Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. JBiomed Sci 27: , 49. |
[16] | Zhang Y , Zhao Y , Liu Y , Wang M , Yu W , Zhang L ((2020) ) Exploring the regulatory roles of circular RNAs in Alzheimer’s disease. Transl Neurodegener 9: , 35. |
[17] | D’Anca M , Fenoglio C , Serpente M , Arosio B , Cesari M , Scarpini EA , Galimberti D ((2019) ) Exosome determinants of physiological aging and age-related neurodegenerative diseases. Front Aging Neurosci 11: , 232. |
[18] | Desai BS , Monahan AJ , Carvey PM , Hendey B ((2007) ) Blood-brain barrier pathology in Alzheimer’s and Parkinson’s disease: Implications for drug therapy. Cell Transplant 16: , 285–299. |
[19] | Lee H , Pienaar IS ((2014) ) Disruption of the blood-brain barrier in Parkinson’s disease: Curse or route to a cure? Front Biosci 19: , 272–280. |
[20] | Gray MT , Woulfe JM ((2015) ) Striatal blood-brain barrier permeability in Parkinson’s disease. J Cereb Blood Flow Metab 35: , 747–750. |
[21] | Morris KV , Mattick JS ((2014) ) The rise of regulatory RNA. Nat Rev Genet 15: , 423–437. |
[22] | Yao RW , Wang Y , Chen LL ((2019) ) Cellular functions of long noncoding RNAs. Nat Cell Biol 21: , 542–551. |
[23] | Lin SP , Ye S , Long Y , Fan Y , Mao HF , Chen MT , Ma QJ ((2016) ) Circular RNA expression alterations are involved in OGD/R-induced neuron injury. Biochem Biophys Res Commun 471: , 52–56. |
[24] | Bartel DP ((2004) ) MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: , 281–297. |
[25] | O’Brien J , Hayder H , Zayed Y , Peng C ((2018) ) Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 9: , 402. |
[26] | Shivdasani RA ((2006) ) MicroRNAs: Regulators of gene expression and cell differentiation. Blood 108: , 3646–3653. |
[27] | Rajman M , Schratt G ((2017) ) MicroRNAs in neural development: From master regulators to fine-tuners. Development 144: , 2310–2322. |
[28] | Sonntag KC ((2010) ) MicroRNAs and deregulated gene expression networks in neurodegeneration. Brain Res 1338: , 48–57. |
[29] | Kim J , Inoue K , Ishii J , Vanti WB , Voronov SV , Murchison E , Hannon G , Abeliovich A ((2007) ) A microRNA feedback circuit in midbrain dopamine neurons. Science 317: , 1220–1224. |
[30] | Schlaudraff F , Gründemann J , Fauler M , Dragicevic E , Hardy J , Liss B ((2014) ) Orchestrated increase of dopamine and PARK mRNAs but not miR-133b in dopamine neurons in Parkinson’s disease. Neurobiol Aging 35: , 2302–2315. |
[31] | Briggs CE , Wang Y , Kong B , Woo TU , Iyer LK , Sonntag KC ((2015) ) Midbrain dopamine neurons in Parkinson’s disease exhibit a dysregulated miRNA and target-gene network. Brain Res 1618: , 111–121. |
[32] | Choi DC , Yoo M , Kabaria S , Junn E ((2018) ) MicroRNA-7 facilitates the degradation of alpha-synuclein and its aggregates by promoting autophagy. Neurosci Lett 678: , 118–123. |
[33] | Liang H , Ding B , Liang J , Shi X , Jiang X , Gao Y ((2018) ) MicroRNA-10a inhibits A30P α-synuclein aggregation and toxicity by targeting proapoptotic protein BCL2L11. Int J Clin Exp Pathol 11: , 624–633. |
[34] | Gandhi PN , Chen SG , Wilson-Delfosse AL ((2009) ) Leucine-rich repeat kinase 2 (LRRK2): A key player in the pathogenesis of Parkinson’s disease. J Neurosci Res 87: , 1283–1295. |
[35] | Cho HJ , Liu G , Jin SM , Parisiadou L , Xie C , Yu J , Sun L , Ma B , Ding J , Vancraenenbroeck R , Lobbestael E , Baekelandt V , Taymans JM , He P , Troncoso JC , Shen Y , Cai H ((2013) ) microRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum Mol Genet 22: , 608–620. |
[36] | Chen Y , Gao C , Sun Q , Pan H , Huang P , Ding J , Chen S ((2017) ) microRNA-4639 is a regulator of DJ-1 expression and a potential early diagnostic marker for Parkinson’s disease. Front Aging Neurosci 9: , 232. |
[37] | Wang H , Ye Y , Zhu Z , Mo L , Lin C , Wang Q , Wang H , Gong X , He X , Lu G , Lu F , Zhang S ((2016) ) miR-124 regulates apoptosis and autophagy process in MPTP model of Parkinson’s disease by targeting to Bim. Brain Pathol 26: , 167–176. |
[38] | Weber JA , Baxter DH , Zhang S , Huang DY , Huang KH , Lee MJ , Galas DJ , Wang K ((2010) ) The microRNA spectrum in 12 body fluids. Clin Chem 56: , 1733–1741. |
[39] | Valadi H , Ekström K , Bossios A , Sjöstrand M , Lee JJ , Lötvall JO ((2007) ) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: , 654–659. |
[40] | Arroyo JD , Chevillet JR , Kroh EM , Ruf IK , Pritchard CC , Gibson DF , Mitchell PS , Bennett CF , Pogosova-Agadjanyan EL , Stirewalt DL , Tait JF , Tewari M ((2011) ) Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 108: , 5003–5008. |
[41] | Magdalinou N , Lees AJ , Zetterberg H ((2014) ) Cerebrospinal fluid biomarkers in parkinsonian conditions: An update and future directions. J Neurol Neurosurg Psychiatry 85: , 1065–1075. |
[42] | Leggio L , Vivarelli S , L’Episcopo F , Tirolo C , Caniglia S , Testa N , Marchetti B , Iraci N ((2017) ) microRNAs in Parkinson’s disease: From pathogenesis to novel diagnostic and therapeutic approaches. Int J Mol Sci 18: , 2698. |
[43] | Roser AE , Caldi Gomes L , Schünemann J , Maass F , Lingor P ((2018) ) Circulating miRNAs as diagnostic biomarkers for Parkinson’s disease. Front Neurosci 12: , 625. |
[44] | Acharya S , Salgado-Somoza A , Stefanizzi FM , Lumley AI , Zhang L , Glaab E , May P , Devaux Y ((2020) ) Non-coding RNAs in the brain-heart axis: The case of Parkinson’s disease. Int J Mol Sci 21: , 6513. |
[45] | Martins M , Rosa A , Guedes LC , Fonseca BV , Gotovac K , Violante S , Mestre T , Coelho M , Rosa MM , Martin ER , Vance JM , Outeiro TF , Wang L , Borovecki F , Ferreira JJ , Oliveira SA ((2011) ) Convergence of miRNA expression profiling, α-synuclein interaction and GWAS in Parkinson’s disease. PLoS One 6: , e25443. |
[46] | Botta-Orfila T , Morató X , Compta Y , Lozano JJ , Falgás N , Valldeoriola F , Pont-Sunyer C , Vilas D , Mengual L , Fernández M , Molinuevo JL , Antonell A , Martí MJ , Fernández-Santiago R , Ezquerra M ((2014) ) Identification of blood serum micro-RNAs associated with idiopathic and LRRK2 Parkinson’s disease. J Neurosci Res 92: , 1071–1077. |
[47] | Ma W , Li Y , Wang C , Xu F , Wang M , Liu Y ((2016) ) Serum miR-221 serves as a biomarker for Parkinson’s disease. Cell Biochem Funct 34: , 511–515. |
[48] | Bai X , Tang Y , Yu M , Wu L , Liu F , Ni J , Wang Z , Wang J , Fei J , Wang W , Huang F , Wang J ((2017) ) Downregulation of blood serum microRNA 29 family in patients with Parkinson’s disease. Sci Rep 7: , 5411. |
[49] | Ozdilek B , Demircan B ((2020) ) Serum microRNA expression levels in Turkish patients with Parkinson’s disease. Int J Neurosci 25: , 1–9. |
[50] | Burgos K , Malenica I , Metpally R , Courtright A , Rakela B , Beach T , Shill H , Adler C , Sabbagh M , Villa S , Tembe W , Craig D , Van Keuren-Jensen K ((2014) ) Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS One 9: , e94839. |
[51] | Gui Y , Liu H , Zhang L , Lv W , Hu X ((2015) ) Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 6: , 37043–37053. |
[52] | Cao XY , Lu JM , Zhao ZQ , Li MC , Lu T , An XS , Xue LJ ((2017) ) microRNA biomarkers of Parkinson’s disease in serum exosome-like microvesicles. Neurosci Lett 644: , 94–99. |
[53] | Margis R , Margis R , Rieder CR ((2011) ) Identification of blood microRNAs associated to Parkinson’s disease. J Biotechnol 152: , 96–101. |
[54] | Serafin A , Foco L , Blankenburg H , Picard A , Zanigni S , Zanon A , Pramstaller PP , Hicks AA , Schwienbacher C ((2014) ) Identification of a set of endogenous reference genes for miRNA expression studies in Parkinson’s disease blood samples. BMC Res Notes 7: , 715. |
[55] | Vallelunga A , Ragusa M , Di Mauro S , Iannitti T , Pilleri M , Biundo R , Weis L , Di Pietro C , De Iuliis A , Nicoletti A , Zappia M , Purrello M , Antonini A ((2014) ) Identification of circulating microRNAs for the differential diagnosis of Parkinson’s disease and Multiple System Atrophy. Front Cell Neurosci 8: , 156. |
[56] | Marques TM , Kuiperij HB , Bruinsma IB , van Rumund A , Aerts MB , Esselink RAJ , Bloem BR , Verbeek MM ((2017) ) microRNAs in cerebrospinal fluid as potential biomarkers for Parkinson’s disease and multiple system atrophy. Mol Neurobiol 54: , 7736–7745. |
[57] | Patil KS , Basak I , Dalen I , Hoedt E , Lange J , Lunde KA , Liu Y , Tysnes OB , Forsgren L , Aarsland D , Neubert TA , Larsen JP , Alves G , Møller SG ((2019) ) Combinatory microRNA serum signatures as classifiers of Parkinson’s disease. Parkinsonism Relat Disord 64: , 202–210. |
[58] | Oliveira SR , Dionísio PA , Correia Guedes L , Gonçalves N , Coelho M , Rosa MM , Amaral JD , Ferreira JJ , Rodrigues CMP ((2020) ) Circulating inflammatory miRNAs associated with Parkinson’s disease pathophysiology. Biomolecules 10: , 945. |
[59] | Zhang X , Yang R , Hu BL , Lu P , Zhou LL , He ZY , Wu HM , Zhu JH ((2017) ) Reduced circulating levels of miR-433 and miR-133b are potential biomarkers for Parkinson’s disease. Front Cell Neurosci 11: , 170. |
[60] | Ravanidis S , Bougea A , Papagiannakis N , Maniati M , Koros C , Simitsi AM , Bozi M , Pachi I , Stamelou M , Paraskevas GP , Kapaki E , Moraitou M , Michelakakis H , Stefanis L , Doxakis E ((2020) ) Circulating brain-enriched microRNAs for detection and discrimination of idiopathic and genetic Parkinson’s disease. Mov Disord 35: , 457–467. |
[61] | Ito M , Uchino H ((2020) ) Chapter Twenty-Three - Detection and quantification of microRNAs (miRNAs) and high-throughput miRNA profiling. In Translational Epigenetics, Epigenetics Methods, Vol 18, Tollefsbol T, ed. Academic Press, 479–493. |
[62] | Tang YT , Huang YY , Zheng L , Qin SH , Xu XP , An TX , Xu Y , Wu YS , Hu XM , Ping BH , Wang Q ((2017) ) Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int J Mol Med 40: , 834–844. |
[63] | Barile L , Vassalli G ((2017) ) Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol Ther 174: , 63–78. |
[64] | Zhang Y , Liu Y , Liu H , Tang WH ((2019) ) Exosomes: Biogenesis, biologic function. Cell Biosci 9: , 19. |
[65] | Cheng L , Sharples RA , Scicluna BJ , Hill AF ((2014) ) Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles 3: , 23743. |
[66] | Yao YF , Qu MW , Li GC , Zhang FB , Rui HC ((2018) ) Circulating exosomal miRNAs as diagnostic biomarkers in Parkinson’s disease. Eur Rev Med Pharmacol Sci 22: , 5278–5283. |
[67] | Dos Santos MCT , Barreto-Sanz MA , Correia BRS , Bell R , Widnall C , Perez LT , Berteau C , Schulte C , Scheller D , Berg D , Maetzler W , Galante PAF , Nogueira da Costa A ((2018) ) miRNA-based signatures in cerebrospinal fluid as potential diagnostic tools for early stage Parkinson’s disease. Oncotarget 9: , 17455–17465. |
[68] | Fatica A , Bozzoni I ((2014) ) Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet 15: , 7–21. |
[69] | Park HJ , Ji P , Kim S , Xia Z , Rodriguez B , Li L , Su J , Chen K , Masamha CP , Baillat D , Fontes-Garfias CR , Shyu AB , Neilson JR , Wagner EJ , Li W ((2018) ) 3’ UTR shortening represses tumor-suppressor genes in trans by disrupting ceRNA crosstalk. Nat Genet 50: , 783–789. |
[70] | Lee B , Sahoo A , Marchica J , Holzhauser E , Chen X , Li JL , Seki T , Govindarajan SS , Markey FB , Batish M , Lokhande SJ , Zhang S , Ray A , Perera RJ ((2017) ) The long noncoding RNA SPRIGHTLY acts as an intranuclear organizing hub for pre-mRNA molecules. Sci Adv 3: , e1602505. |
[71] | Statello L , Guo CJ , Chen LL , Huarte M ((2021) ) Gene regulation by long non-coding RNAs and its biological functions. Cell Biol 22: , 96–118. |
[72] | Wei CW , Luo T , Zou SS , Wu AS ((2018) ) The role of long noncoding RNAs in central nervous system and neurodegenerative diseases. Front Behav Neurosci 12: , 175. |
[73] | Li L , Zhuang Y , Zhao X , Li X ((2018) ) Long non-coding RNA in neuronal development and neurological disorders. Front Genet 9: , 744. |
[74] | Andersen RE , Lim DA ((2017) ) Forging our understanding of lncRNAs in the brain. Cell Tissue Res 371: , 55–71. |
[75] | Majidinia M , Mihanfar A , Rahbarghazi R , Nourazarian A , Bagca B , Avci ÇB ((2016) ) The roles of non-coding RNAs in Parkinson’s disease. Mol Biol Rep 43: , 1193–1204. |
[76] | Kraus TFJ , Haider M , Spanner J , Steinmaurer M , Dietinger V , Kretzschmar HA ((2017) ) Altered long noncoding RNA expression precedes the course of Parkinson’s disease-a preliminary report. Mol Neurobiol 54: , 2869–2877. |
[77] | Boros FA , Maszlag-Török R , Vécsei L , Klivényi P ((2020) ) Increased level of NEAT1 long non-coding RNA is detectable in peripheral blood cells of patients with Parkinson’s disease. Brain Res 1730: , 146672. |
[78] | Ni Y , Huang H , Chen Y , Cao M , Zhou H , Zhang Y ((2017) ) Investigation of long non-coding RNA expression profiles in the substantia nigra of Parkinson’s disease. Cell Mol Neurobiol 37: , 329–333. |
[79] | Lv Q , Wang Z , Zhong Z , Huang W ((2020) ) Role of long noncoding RNAs in Parkinson’s disease: Putative biomarkers and therapeutic targets. Parkinsons Dis 2020: , 5374307. |
[80] | Elkouris M , Kouroupi G , Vourvoukelis A , Papagiannakis N , Kaltezioti V , Matsas R , Stefanis L , Xilouri M , Politis PK ((2019) ) Long non-coding RNAs associated with neurodegeneration-linked genes are reduced in Parkinson’s disease patients. Front Cell Neurosci 13: , 58. |
[81] | Scheele C , Petrovic N , Faghihi MA , Lassmann T , Fredriksson K , Rooyackers O , Wahlestedt C , Good L , Timmons JA ((2007) ) The human PINK1 locus is regulatedby a non-coding natural antisense RNA during modulation of mitochondrial function. BMC Genomics 8: , 74. |
[82] | Carrieri C , Forrest AR , Santoro C , Persichetti F , Carninci P , Zucchelli S , Gustincich S ((2015) ) Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front Cell Neurosci 9: , 114. |
[83] | Coupland KG , Kim WS , Halliday GM , Hallupp M , Dobson-Stone C , Kwok JB ((2016) ) Role of the long non-coding RNA MAPT-AS1 in regulation of Microtubule Associated Protein Tau (MAPT) expression in Parkinson’s disease. PLoS One 11: , e0157924. |
[84] | Soreq L , Guffanti A , Salomonis N , Simchovitz A , Israel Z , Bergman H , Soreq H ((2014) ) Long non-coding RNA and alternative splicing modulations in Parkinson’s leukocytes identified by RNA sequencing. PLoS Comput Biol 10: , e1003517. |
[85] | Zhou Y , Gu C , Li J , Zhu L , Huang G , Dai J , Huang H ((2018) ) Aberrantly expressed long noncoding RNAs and genes in Parkinson’s disease. Neuropsychiatr Dis Treat 14: , 3219–3229. |
[86] | Chi LM , Wang LP , Jiao D ((2019) ) Identification of differentially expressed genes and long noncoding RNAs associated with Parkinson’s disease. Parkinsons Dis 2019: , 6078251. |
[87] | Wang Q , Han CL , Wang KL , Sui YP , Li ZB , Chen N , Fan SY , Shimabukuro M , Wang F , Meng FG ((2020) ) Integrated analysis of exosomal lncRNA and mRNA expression profiles reveals the involvement of lnc-MKRN2-42:1 In the pathogenesis of Parkinson’s disease. CNS Neurosci Ther 26: , 527–537. |
[88] | Fan Y , Li J , Yang Q , Gong C , Gao H , Mao Z , Yuan X , Zhu S , Xue Z ((2019) ) Dysregulated long non-coding RNAs in Parkinson’s disease contribute to the apoptosis of human neuroblastoma cells. Front Neurosci 13: , 1320. |
[89] | Xilouri M , Brekk OR , Stefanis L ((2013) ) α-Synuclein and protein degradation systems: A reciprocal relationship. Mol Neurobiol 47: , 537–551. |
[90] | Carrieri C , Cimatti L , Biagioli M , Beugnet A , Zucchelli S , Fedele S , Pesce E , Ferrer I , Collavin L , Santoro C , Forrest AR , Carninci P , Biffo S , Stupka E , Gustincich S ((2012) ) Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491: , 454–457. |
[91] | Bhattacharjee N , Borah A ((2016) ) Oxidative stress and mitochondrial dysfunction are the underlying events of dopaminergic neurodegeneration in homocysteine rat model of Parkinson’s disease. Neurochem Int 10: , 48–55. |
[92] | Simchovitz A , Hanan M , Niederhoffer N , Madrer N , Yayon N , Bennett ER , Greenberg DS , Kadener S , Soreq H ((2019) ) NEAT1 is overexpressed in Parkinson’s disease substantia nigra and confers drug-inducible neuroprotection from oxidative stress. FASEB J 33: , 11223–11234. |
[93] | Chen Q , Huang X , Li R ((2018) ) lncRNA MALAT1/miR-205-5p axis regulates MPP+-induced cell apoptosis in MN9D cells by directly targeting LRRK2. Am J Transl Res 10: , 563–572. |
[94] | Wilusz JE ((2017) ) Circular RNAs: Unexpected outputs of many protein-coding genes. RNA Biol 14: , 1007–1017. |
[95] | Hansen TB , Jensen TI , Clausen BH , Bramsen JB , Finsen B , Damgaard CK , Kjems J ((2013) ) Natural RNA circles function as efficient microRNA sponges. Nature 495: , 384–388. |
[96] | Li Z , Huang C , Bao C , Chen L , Lin M , Wang X , Zhong G , Yu B , Hu W , Dai L , Zhu P , Chang Z , Wu Q , Zhao Y , Jia Y , Xu P , Liu H , Shan G ((2015) ) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22: , 256–264. |
[97] | Memczak S , Jens M , Elefsinioti A , Torti F , Krueger J , Rybak A , Maier L , Mackowiak SD , Gregersen LH , Munschauer M , Loewer A , Ziebold U , Landthaler M , Kocks C , le Noble F , Rajewsky N ((2013) ) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: , 333–338. |
[98] | Li TR , Jia YJ , Wang Q , Shao XQ , Lv RJ ((2017) ) Circular RNA: A new star in neurological diseases. Int J Neurosci 127: , 726–734. |
[99] | Qu S , Yang X , Li X , Wang J , Gao Y , Shang R , Sun W , Dou K , Li H ((2015) ) Circular RNA: A new star of noncoding RNAs. Cancer Lett 365: , 141–148. |
[100] | Piwecka M , Glažar P , Hernandez-Miranda LR , Memczak S , Wolf SA , Rybak-Wolf A , Filipchyk A , Klironomos F , Cerda Jara CA , Fenske P , Trimbuch T , Zywitza V , Plass M , Schreyer L , Ayoub S , Kocks C , Kühn R , Rosenmund C , Birchmeier C , Rajewsky N ((2017) ) Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357: , eaam8526. |
[101] | Li Y , Zheng Q , Bao C , Li S , Guo W , Zhao J , Chen D , Gu J , He X , Huang S ((2015) ) Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res 25: , 981–984. |
[102] | Mehta SL , Dempsey RJ , Vemuganti R ((2020) ) Role of circular RNAs in brain development and CNS diseases. Prog Neurobiol 186: , 101746. |
[103] | Lukiw WJ ((2013) ) Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet 4: , 307. |
[104] | Hanan M , Simchovitz A , Yayon N , Vaknine S , Cohen-Fultheim R , Karmon M , Madrer N , Rohrlich TM , Maman M , Bennett ER , Greenberg DS , Meshorer E , Levanon EY , Soreq H , Kadener S ((2020) ) A Parkinson’s disease CircRNAs resource reveals a link between circSLC8A1 and oxidative stress. EMBO Mol Med 12: , e11942. |
[105] | Tan CL , Plotkin JL , Venø MT , von Schimmelmann M , Feinberg P , Mann S , Handler A , Kjems J , Surmeier DJ , O’Carroll D , Greengard P , Schaefer A ((2013) ) microRNA-128 governs neuronal excitability and motor behavior in mice. Science 342: , 1254–1258. |
[106] | Sang Q , Liu X , Wang L , Qi L , Sun W , Wang W , Sun Y , Zhang H ((2018) ) CircSNCA downregulation by pramipexole treatment mediates cell apoptosis and autophagy in Parkinson’s disease by targeting miR-7. Aging (Albany NY) 10: , 1281–1293. |
[107] | Jia E , Zhou Y , Liu Z , Wang L , Ouyang T , Pan M , Bai Y , Ge Q ((2020) ) Transcriptomic profiling of circular RNA in different brain regions of Parkinson’s disease in a mouse model. Int J Mol Sci 21: , 3006. |
[108] | Maass PG , Glazar P , Memczak S , Dittmar G , Hollfinger I , Schreyer L , Sauer AV , Toka O , Aiuti A , Luft FC , Rajewsky N ((2017) ) . A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl) 95: , 1179–1189. |
[109] | Memczak S , Papavasileiou P , Peters O , Rajewsky N ((2015) ) Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One 10: , e0141214. |
[110] | Lee S , Mankhong S , Kang JH ((2019) ) Extracellular vesicle as a source of Alzheimer’s biomarkers: Opportunities and challenges. Int J Mol Sci 20: , 1728. |
[111] | Pinho R , Guedes LC , Soreq L , Lobo PP , Mestre T , Coelho M , Rosa MM , Gonçalves N , Wales P , Mendes T , Gerhardt E , Fahlbusch C , Bonifati V , Bonin M , Miltenberger-Miltényi G , Borovecki F , Soreq H , Ferreira JJ , Outeiro F T ((2016) ) Gene expression differences in peripheral blood of Parkinson’s disease patients with distinct progression profiles. PLoS One 11: , e0157852. |
[112] | Soreq L , Bergman H , Israel Z , Soreq H ((2012) ) Exon arrays reveal alternative splicing aberrations in Parkinson’s disease leukocytes. Neurodegener Dis 10: , 203–206. |
[113] | Soreq L , Salomonis N , Bronstein M , Greenberg DS , Israel Z , Bergman H , Soreq H ((2013) ) Small RNA sequencing-microarray analyses in Parkinson leukocytes reveal deep brain stimulation-induced splicing changes that classify brain region transcriptomes. Front Mol Neurosci 6: , 10. |
[114] | Ravanidis S , Bougea A , Karampatsi D , Papagiannakis N , Maniati M , Stefanis L , Doxakis E ((2021) ) Differentially expressed circular RNAs in peripheral blood mononuclear cells of patients with Parkinson’s disease. Mov Disord 36: , 1170–1179. |
[115] | Salta E , De Strooper B ((2012) ) Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol 11: , 189–200. |
[116] | Xin C , Liu J ((2021) ) Long non-coding RNAs in Parkinson’s disease. Neurochem Res 46: , 1031–1042. |
[117] | Sweeney MD , Sagare AP , Zlokovic BV ((2018) ) Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat Rev Neurol 14: , 133–150. |




