Identification of Full-Length Wild-Type and Mutant Huntingtin Interacting Proteins by Crosslinking Immunoprecipitation in Mice Brain Cortex
Abstract
Background:
Huntington’s disease is a neurodegenerative disorder caused by a CAG expansion in the huntingtin gene, resulting in a polyglutamine expansion in the ubiquitously expressed mutant huntingtin protein.
Objective:
Here we set out to identify proteins interacting with the full-length wild-type and mutant huntingtin protein in the mice cortex brain region to understand affected biological processes in Huntington’s disease pathology.
Methods:
Full-length huntingtin with 20 and 140 polyQ repeats were formaldehyde-crosslinked and isolated via their N-terminal Flag-tag from 2-month-old mice brain cortex. Interacting proteins were identified and quantified by label-free liquid chromatography-mass spectrometry (LC-MS/MS).
Results:
We identified 30 interactors specific for wild-type huntingtin, 14 interactors specific for mutant huntingtin and 14 shared interactors that interacted with both wild-type and mutant huntingtin, including known interactors such as F8a1/Hap40. Syt1, Ykt6, and Snap47, involved in vesicle transport and exocytosis, were among the proteins that interacted specifically with wild-type huntingtin. Various other proteins involved in energy metabolism and mitochondria were also found to associate predominantly with wild-type huntingtin, whereas mutant huntingtin interacted with proteins involved in translation including Mapk3, Eif3h and Eef1a2.
Conclusion:
Here we identified both shared and specific interactors of wild-type and mutant huntingtin, which are involved in different biological processes including exocytosis, vesicle transport, translation and metabolism. These findings contribute to the understanding of the roles that wild-type and mutant huntingtin play in a variety of cellular processes both in healthy conditions and Huntington’s disease pathology.
INTRODUCTION
Huntington’s disease (HD) is caused by a CAG-repeat expansion in exon1 of the huntingtin (Htt) gene, the inheritance is autosomal dominant and the course of the disease is progressive. HD occurs in individuals whose Htt gene contains 40 or more CAG repeats [1], with longer repeat lengths generally resulting in an earlier age of onset [2]. The resulting polyglutamine (polyQ) expansion is thought to cause conformational changes in the mutant Htt protein, leading to altered post-translational modifications and protein-protein interactions, and finally neuronal cell death. HD associated symptoms include motor impairment including chorea along with psychiatric and cognitive dysfunctions that progress to death over 15–25 years [3]. General brain atrophy, inclusion body formation in central nervous system neurons, a drastic loss of medium spiny neurons in the striatum, and loss of neurons in the deeper layer of the cortex are neuropathological features of the disorder [4].
Htt is a 348 kDa protein that is essential for embryogenesis. The Htt protein contains repetitive structural components with scaffolding properties called HEAT-repeats [5]. Huntingtin can interact with numerous organelles, such as the endoplasmatic reticulum, Golgi complex, and mitochondria [6, 7]. Additionally, Htt was found to be associated with vesicular structures such as clathrin-coated vesicles, endosomal compartments or caveolae, and microtubules in neurites and synapses [8]. Several protein-protein interaction studies have suggested roles for Htt in cellular processes including endocytosis, vesicle transport, post-synaptic signaling, calcium homeostasis and gene regulation [9]. The goal of our study was to identify Htt interacting proteins in order to better understand the functions of wild-type huntingtin (wtHtt) and mutant huntingtin (mHtt) by using formaldehyde crosslinking in order to stabilize potential transient protein complexes. Formaldehyde forms covalent bonds between proteins which are in close proximity (2.3 –2.7 Å). Importantly, formaldehyde crosslinks are reversible and compatible with mass spectrometry [10]. Here, we immunoprecipitated (IP) Flag-tagged wtHtt and mHtt from mice brain cortex from established knock-in HD mice models, expressing N-terminal 3xFlag epitope tagged full-length (fl) Htt with Q20 or Q140 polyglutamine stretches [11]. As a negative control we used non-Flag-tagged wtHtt mice. In order to identify proteins interacting with soluble Htt we used young mice (2-month-old) since mutant Htt mice start developing aggregates around 6 months of age [11]. Htt interacting proteins were de-crosslinked and subsequently identified by mass spectrometry-based label-free quantification (LFQ).
MATERIAL AND METHODS
Brain tissue sample preparation for formaldehyde crosslinking immunoprecipitations
Snap frozen left and right cortices from three different groups of 2-month-old mice were used. The wild-type group, denoted as Q20, were wtHtt mice expressing 3xFlag-tagged fl Htt with 20 polyQ repeats. The mutant group, denoted as Q140, were mHtt knock-in mice expressing 3xFlag-tagged fl Htt with 140 polyQ repeats. Both Q20 and Q140 samples were provided by Scott Zeitlin [11, 12] and contain a chimeric mouse/human exon1. Finally, the negative control group were wtHtt mice expressing Htt without the Flag-tag. Biological quadruplicates, each containing three cortices, were analyzed. Percentage of formaldehyde was optimized for our samples by resolving crosslinked (boiled in SDS-PAGE buffer for 5 min at 65°C) and de-crosslinked (boiled in SDS-PAGE sample buffer for 20 min at 99°C) wild-type mice brain lysates on SDS-PAGE and by checking for high molecular weight complex formation of both tubulin and Htt with different formaldehyde concentrations (Supplementary Figure 1). Best crosslinking results were obtained with 0.5%formaldehyde. Frozen cortices were grinded and crosslinked by shaking 10 min in 0.5%formaldehyde at room temperature. Subsequently the formaldehyde was removed and the reaction was quenched with 250 mM Tris pH 8.0 for 1 min at room temperature. Tris was removed and lysis buffer (50 mM Tris/HCl pH 7.4; 150 mM NaCl; 1 mM EDTA; 1%Triton X-100; Roche protease inhibitor tablets (Roche Diagnostics GmbH, Mannheim, Germany)) was added in 1:5 ratio of the volume of grinded brain material. Samples were sonicated in 4 rounds of 5 cycles of 40 s on, 20 s off at amplitude 40 (Q800R3, QSonica) and centrifuged for 30 min at 14 krpm at 4°C, after which the supernatant was transferred to a new tube. Pellet fraction was washed with 50μl lysis buffer, centrifuged for 5 min at 14 krpm and this was added to the supernatant. Protein concentration was determined by Bradford assay.
Immunoprecipitation (IP)
Antibody coupling
Antibodies were coupled to the beads as described earlier [13]. Briefly, 2 mg α-Flag M2 antibodies (#SLBV9325, Sigma-Aldrich) were coupled to 1 ml ProtG beads (#17061801, GE Healthcare) by using dimethyl pimelimidate. Coupling was verified by SDS-PAGE and Coomassie staining.
IP and formaldehyde de-crosslinking
Ten milligrams of protein were used per IP. Lysates were precleared by rotating 30 min with 100μl empty ProtG beads at 4°C. Precleared lysates were incubated O/N with 100μl α-FLAG-coupled beads in a rotator at 4°C. Beads were washed 3x with wash buffer I (50 mM Tris pH 7.4; 300 mM NaCl; 1 mM EDTA; 0.1%NP40) and washed 1x with wash buffer II (50 mM Tris pH 7.4; 150 mM NaCl; 1 mM EDTA; 0.01%NP40). Samples were eluted by boiling in sample loading buffer. Samples were boiled for 20 min at 99°C to break formaldehyde crosslinks. Proteins were resolved by 4–15%precast SDS-PAGE gradient gels (#456–1084, BioRad) and visualized by Coomassie staining.
Sample preparation for mass spectrometry
SDS-PAGE gel lanes were cut in 1 mm slices and combined into a total of 7 fractions per lane. In-gel protein reduction, alkylation and tryptic digestion were performed as described previously [14]. Peptides were extracted with 30%acetonitrile/0.5%formic acid and analyzed on an EASY-nLC 1000 LC system (ThermoFisher Scientific) coupled to an Orbitrap Fusion™ Tribrid™ Mass Spectrometer (ThermoFisher Scientific). Peptide mixtures were trapped on an Acclaim™ PepMap™ 100 C18 reversed phase column (100μm x 2 cm, 5μm beads, pore size 100 Å, ThermoFisher Scientific) at a flow rate of 1μl/min. Peptide separation was performed on a 100μm x 20 cm reversed phase column packed in-house with 2.5μm BEH C18 particles (Waters) and using a 60-min gradient from 0%B (= 100%A) to 50%B (A = 0.1%formic acid; B = 80%(v/v) acetonitrile, 0.1%formic acid) at a constant flow rate of 500 nl/min. The column eluent was directly electrosprayed into the mass spectrometer. MS1 spectra were acquired in continuum mode; fragmentation of the peptides was performed in data-dependent acquisition mode by HCD at 30%collision energy using 3 s cycle time and a 30 s dynamic exclusion filter and 1.6 m/z isolation window. Additional settings: MS1 orbitrap resolution 60K; MS1 AGC target 5E5; MS2 AGC target 7E3; MS1 and MS2 maximum injection time 50 ms; MS1 scan range 375–1500 m/z; MS1 intensity threshold 1E4; MS2 detection scan rate in the ion trap was set at rapid.
Mass spectrometry data analysis
The raw files were analyzed using MaxQuant [15] software package (v1.6.5.0). The Swiss-Prot Mus musculus proteome downloaded from UniProt (June 2019, 17,010 entries) together with the revert decoys and standards contaminants database from MaxQuant were used in the search. Trypsin was selected as an enzyme with maximum number of missed cleavages allowed up to 3. Methionine oxidation and N-terminal acetylation were selected as variable modifications, carbamidomethylation of cysteine was selected as a fixed modification. Maximum modifications allowed per peptide was set to 5. The default MaxQuant settings were used for precursor tolerance (20 ppm for the first search, 4.5 ppm for the main search) and fragment ion tolerance. The false discovery rates for peptide spectrum matches and protein identifications were set to 1%.
The MaxQuant output had 1,838 identifications in total for control, Q20 and Q140. Statistical analysis was done using an in-house R-script. For each identification, the first protein listed in the proteinGroups was selected as the identified protein. Then the proteins that were identified by only one peptide, only identified by post-translational modification site, marked as a reverse sequence or as a potential contaminant were filtered out leaving a total number of 1,133 identifications. Intensity values were transformed to log2 values. The missing values were imputed by taking a random value from the 1st quartile of the whole distribution of log2 values, excluding the NA measurements. Supplementary Table 1 contains the data before and after imputation. Proteins identified in at least 3 out of the 4 replicates in Q20 or Q140 datasets were compared with the control. Q20 vs. control table had 1,074 proteins and the Q140 vs. control table had 991 proteins for comparison. Two-sided, unpaired t-test was done to compare the proteins in each table. A p-value cut-off of < 0.05 and a log2 fold change cut-off of > 1 was used to select for significantly upregulated proteins in the Q-lengths. In the end, 44 and 28 significantly abundant proteins were discovered in Q20 vs. control and Q140 vs. control, respectively (Supplementary Table 2).
GO term enrichment
GOrilla web-based tool [16] was used to perform GO enrichment analysis on proteins upregulated in Q20 and Q140. Upregulated proteins were used as the target set and remaining proteins identified in at least 3 out of 4 replicates were used as the background set. p-value threshold was 0.001. Results are given in Supplementary Table 3. Significantly enriched biological processes were summarized and visualized using REVIGO [17] with SimRel score as the semantic similarity score and allowed similarity set to medium (Fig. 2E).
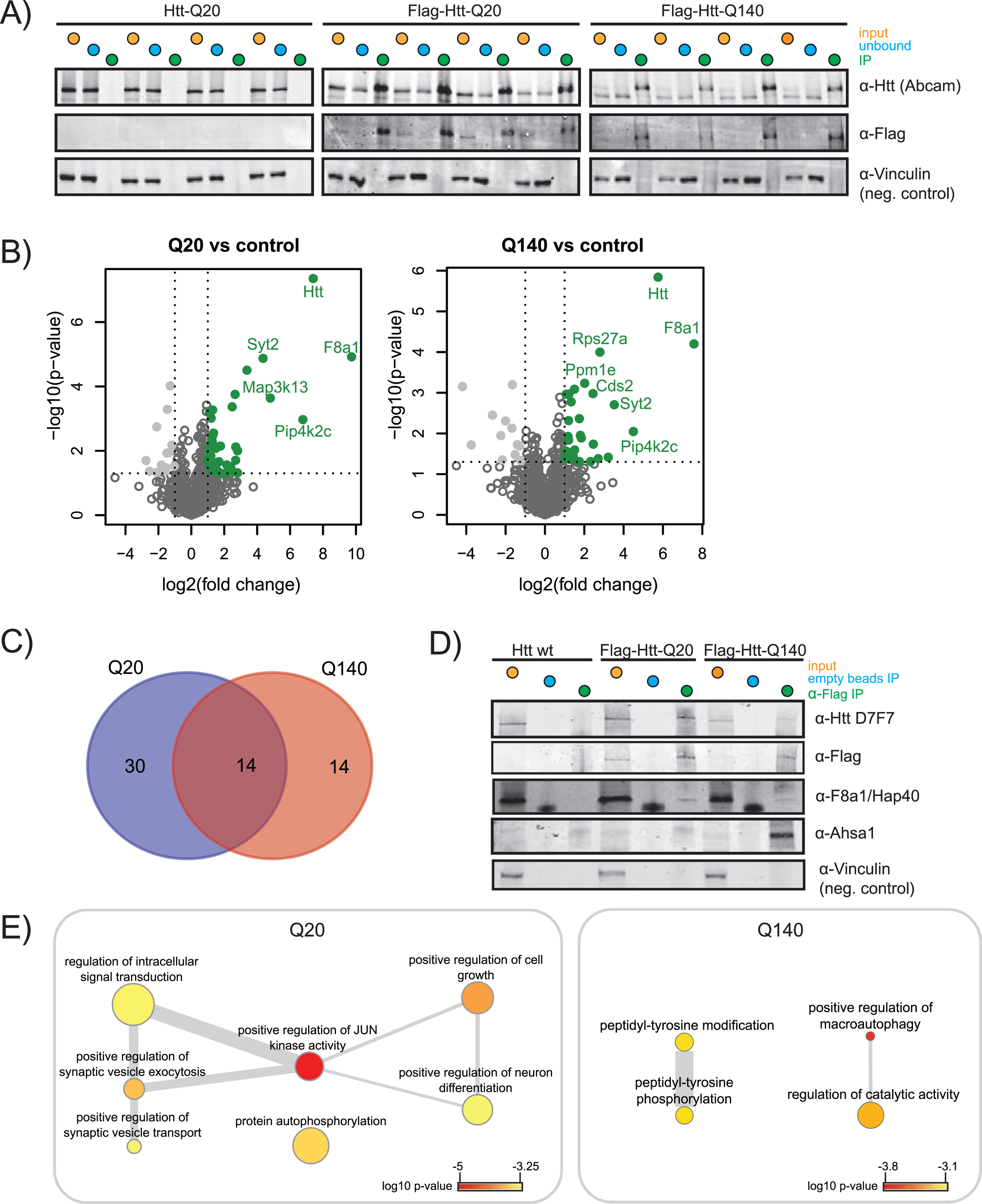
Fig. 2
Immunoprecipitation of wtHtt and mHtt interacting proteins. A) SDS-PAGE western blot (WB) of input, unbound fraction and immunoprecipitation of quadruplicate control (Htt-Q20), wild-type Htt (Flag-Htt-Q20) and mutant Htt (Flag-Htt-Q140) samples. WB was performed using antibodies directed against the Htt N-terminus, Flag-tag, and an antibody directed against vinculin was used as a negative control. B) Volcano plots showing proteins significantly up in Q20 vs. control and Q140 vs. control. The proteins that pass the p-value and log2 fold change threshold are shown in green. The tables for Q-length groups vs. control are given in the Supplementary Table 2. C) Venn diagram showing the overlap between the proteins significantly up in Q20 vs. control and Q140 vs. control. D) SDS-PAGE WB was performed to validate the results obtained by mass spectrometry. Specific validation of Flag-tagged wtHtt and mHtt was done using D7F7 antibody for Htt and α-Flag antibody. Validation of wtHtt and mHtt interactor F8a1/Hap40 and mHtt interactor Ahsa1 are shown. Vinculin was used as a negative control. E) Biological processes enriched in Q20 and Q140. p-value indicated by color, frequency of GO-term indicated by node size, degree of similarity indicated by thickness of edges.
Pathway analysis
Protein-protein association networks for proteins that co-precipitated significantly with Htt Q20 or Htt Q140 were visualized using the StringApp [18] (v1.6.0) in Cytoscape [19] (v.3.8.2). Cytoscape plugin Molecular Complex Detection (MCODE) [20] (v2.0.0) with default settings (degree cutoff: 2; cluster finding: haircut; Node Score Cutoff: 0.2; K-Core: 2; Max Depth: 100) was used to identify highly interconnected clusters.
SDS-PAGE and western blot analysis
Antibodies used for SDS-PAGE western blot (WB) analysis: α-Htt (ab109115, Abcam); α-Htt D7F7 (#5656 CST); α-Flag M2 (#SLBV9325, Sigma-Aldrich); α-Vinculin (#V9131, Sigma-Aldrich); α-F8A1/Hap40 (#PA5-61382, Invitrogen), α-AHSA1 (#12841, CST). SDS-PAGE WB was performed according to standard procedures, except for Htt (∼350kDa). For Htt a 6%separating tris-glycine SDS-PAGE gel was used and proteins were run out of the gel until ∼80 kDa. Wet blotting was done O/N at 45V at 4°C. Proteins were visualized using LiCOR secondary antibodies and the Odyssey system.
RESULTS
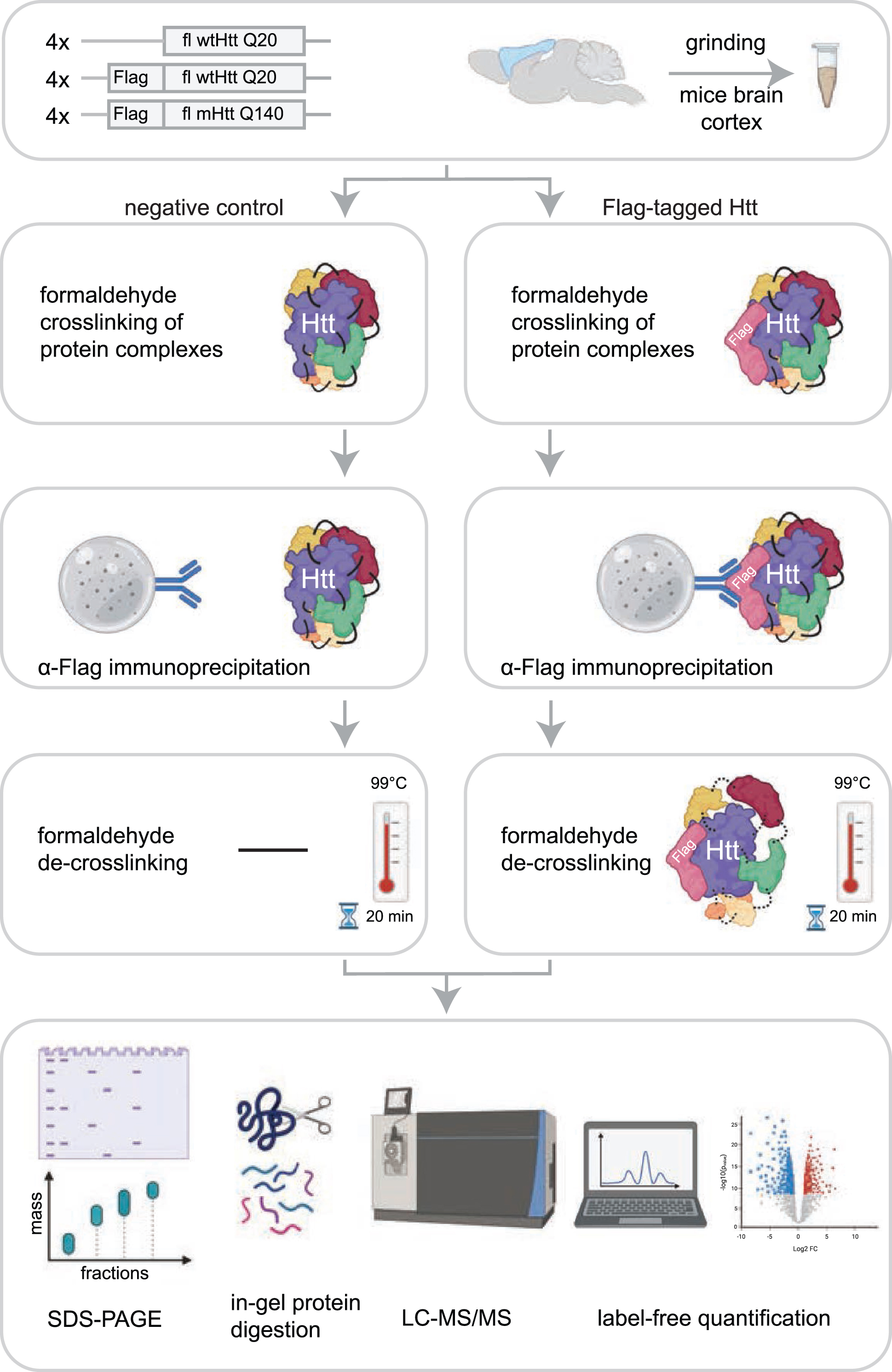
In this study, we set out to identify novel interactors of fl wtHtt and mHtt in mice brain cortex material. In order to identify additional proteins as compared to other protein-protein interaction studies performed in HD models, we used formaldehyde crosslinking, which stabilizes transient protein complexes. This treatment is reversible and is compatible with mass spectrometry. To focus on protein interactions with soluble fl Htt, we used Htt Q20 and Q140 knock-in mice that were two months old, since at this age the used HD model does not contain aggregates (Supplementary Figure 2) [11]. The mice expressed heterozygous N-terminal Flag-tagged fl Htt with either 20 polyglutamine repeats (wtHtt), or 140 polyglutamine repeats (mHtt). The control mice expressed endogenous fl wtHtt without the Flag-tag. All samples were prepared in biological quadruplicates (Fig. 1). To stabilize transient interactions, protein complexes were crosslinked by incubation with 0.5%formaldehyde for 10 min. IP of the crosslinked material was performed via protG-coupled anti-Flag antibodies, after which the precipitated proteins were de-crosslinked by boiling for 20 min in sample loading buffer. Next, the proteins were resolved by SDS-PAGE, digested in-gel using trypsin, and analyzed by liquid chromatography-mass spectrometry (LC-MS/MS). Relative protein abundances of the negative control were compared with the samples containing Flag-tagged Htt using Label-Free Quantification (Fig. 1).
Fig. 1
Workflow for identification of Htt interactors via crosslinking immunoprecipitation and mass spectrometry. The cortex brain regions of 2-month-old mice expressing fl wild-type Htt (wtHtt), Flag-tagged wtHtt and Flag-tagged mutant Htt (mHtt) were used in this study. Four biological replicates were analyzed for each sample. Protein complexes were crosslinked in grinded brain material using 0.5%formaldehyde and were subsequently enriched from lysates using α-Flag coupled beads. After several washing steps the protein complexes were de-crosslinked by boiling for 20 min at 99°C in sample loading buffer. Next, proteins were resolved by SDS-PAGE and analyzed using mass spectrometry-based label-free quantification.
Performance of the IPs was validated by SDS-PAGE WB and shows significant precipitation of Flag-tagged wtHtt and mHtt, while the non-Flag-tagged negative control showed no Htt precipitation (indicated as IP, Fig. 2A). The label-free quantification algorithm of MaxQuant, MaxLFQ, was used for quantification of the proteins identified in LC-MS/MS. The relative fold changes of proteins identified in the Flag-tagged Htt samples compared to the non-Flag-tagged negative controls are shown in the volcano plots (Fig. 2B). We identified 44 putative wtHtt interactors and 28 putative mHtt interactors, shown on the upper-right quadrant (green, in color) of the volcano plots. F8a1/Hap40 was relatively the most abundant interactor of both wtHtt and mHtt. We identified 14 putative interactors that were shared between both forms of Htt (Fig. 2C). Table 1 gives an overview of shared and unique wtHtt and mHtt interactors. Almost 1/3 of the proteins that we identified as an interactor of wtHtt and/or mHtt were identified as Htt interactors in previous studies as well (Table 1). We validated the expression and precipitation of Htt by SDS-PAGE WB. The D7F7 antibody recognizes the residues surrounding Pro1220 of the human huntingtin protein. As expected, this antibody detected Htt in all input samples including endogenous Htt in the negative control, while it only detected Htt in the IP samples of the Flag-tagged brain samples (Fig. 2D). The α-Flag antibody did not detect Htt in the negative control, as expected. Mass spectrometry analysis revealed that Hap40 interacted with both wtHtt and mHtt, while Ahsa1 was a specific interactor of mHtt (Table 1), and we could confirm these results in formaldehyde crosslinked mice brain cortex samples by SDS-PAGE WB (Fig. 2D). In addition, an IP directed against the Flag-tag was performed in non-crosslinked mice cortex lysates. Hap40 was precipitated in both wtHtt and mHtt lysates while Ahsa1 was not, suggesting that Hap40 is a stable interactor whereas Ahsa1 might be a transient interactor of mHtt (Supplementary Figure 3). Next, Gene Ontology (GO) enrichment analysis was performed on the significant interactors of wtHtt and mHtt (Fig. 2E). Biological processes enriched in wtHtt interactors include positive regulation of JUN kinase activity, positive regulation of synaptic vesicle exocytosis and transport, protein autophosphorylation and positive regulation of neuron differentiation. Biological processes enriched in mHtt interactors include peptidyl-tyrosine phosphorylation and positive regulation of macroautophagy (Fig. 2E, Supplementary Table 3).
Table 1
Significantly precipitated proteins in Htt Q20 and Htt Q140 IPs compared to negative control1
| Uniprot | Gene | Log2 fc Q20 | p-value Q20 | Log2 fc Q140 | p-value Q140 | Previously identified |
| Q5SSL4 | Abr | 1.68 | 2.2E-02 | 1.44 | 3.7E-02 | |
| Q99L43 | Cds2 | 3.38 | 3.1E-05 | 2.44 | 1.1E-03 | |
| Q03137 | Epha4 | 1.30 | 3.3E-03 | 1.33 | 1.7E-03 | |
| Q00558 | F8a1 (Hap40) | 9.73 | 1.2E-05 | 7.58 | 6.3E-05 | [43] |
| Q8C0C7 | Farsa | 1.18 | 1.4E-02 | 1.50 | 8.2E-04 | |
| Q6NS60 | Fbxo41 | 1.23 | 4.5E-03 | 1.20 | 4.8E-03 | |
| P42859 | Htt | 7.40 | 4.4E-08 | 5.75 | 1.5E-06 | |
| Q8BKC5 | Ipo5 | 2.81 | 1.0E-02 | 2.46 | 1.8E-02 | |
| P28740 | Kif2a | 1.77 | 7.1E-03 | 1.85 | 1.3E-02 | [43] |
| P28738 | Kif5c | 1.20 | 9.7E-04 | 1.08 | 1.1E-03 | [43] |
| Q91XU3 | Pip4k2c | 6.78 | 1.1E-03 | 4.50 | 9.0E-03 | |
| Q9QVP9 | Ptk2b | 1.49 | 8.3E-03 | 1.19 | 3.7E-02 | [43] |
| P46097 | Syt2 | 4.35 | 1.3E-05 | 3.53 | 2.0E-03 | [43] |
| Q9D6F9 | Tubb4a | 1.11 | 1.8E-02 | 1.22 | 1.1E-03 | |
| Q91V92 | Acly | 1.10 | 3.2E-02 | [43] | ||
| Q60875 | Arhgef2 | 1.09 | 6.3E-04 | [43] | ||
| P56382 | Atp5f1e | 2.81 | 4.8E-02 | |||
| Q78IK2 | Atp5md | 1.41 | 2.4E-02 | |||
| Q9CXW3 | Cacybp | 1.17 | 3.5E-02 | |||
| Q6PHZ2 | Camk2d | 1.53 | 2.3E-02 | |||
| Q8BI72 | Cdkn2aip | 1.01 | 2.0E-02 | |||
| Q9QZS0 | Col4a3 | 2.16 | 2.7E-02 | |||
| Q60737 | Csnk2a1 | 2.55 | 4.6E-02 | [43, 44] | ||
| O88712 | Ctbp1 | 1.11 | 8.8E-03 | [43, 52] | ||
| Q8K1M6 | Dnm1l | 1.55 | 4.0E-02 | |||
| P10630 | Eif4a2 | 1.37 | 3.7E-02 | |||
| Q8VE33 | Gdap1l1 | 1.05 | 8.6E-03 | |||
| Q68FF6 | Git1 | 2.39 | 3.2E-02 | [53] | ||
| O70325 | Gpx4 | 2.69 | 2.0E-02 | |||
| P10922 | H1f0 | 2.70 | 7.6E-03 | |||
| Q9EQ06 | Hsd17b11 | 2.48 | 4.3E-04 | |||
| Q8BNW9 | Kbtbd11 | 1.29 | 5.4E-04 | [43] | ||
| Q60700 | Map3k12 | 2.66 | 1.8E-04 | |||
| Q1HKZ5 | Map3k13 | 4.80 | 2.3E-04 | |||
| Q9QYF9 | Ndrg3 | 1.27 | 5.2E-03 | |||
| Q6PAK3 | Prmt8 | 1.02 | 2.1E-02 | |||
| Q2PFD7 | Psd3 | 1.28 | 5.8E-03 | |||
| Q8CHG7 | Rapgef2 | 1.53 | 4.5E-02 | |||
| Q9Z268 | Rasal1 | 1.24 | 3.6E-02 | [43] | ||
| Q9Z2Z6 | Slc25a20 | 1.31 | 4.1E-02 | |||
| Q8R570 | Snap47 | 1.61 | 4.8E-02 | |||
| P46096 | Syt1 | 1.35 | 2.8E-03 | [43] | ||
| Q9R1R2 | Trim3 | 1.70 | 4.7E-02 | [43] | ||
| Q9CQW1 | Ykt6 | 2.08 | 4.9E-02 | |||
| Q8BK64 | Ahsa1 | 1.27 | 4.3E-02 | [43, 45] | ||
| O54774 | Ap3d1 | 1.17 | 1.2E-02 | [45] | ||
| Q9CPW0 | Cntnap2 | 1.75 | 4.9E-02 | |||
| Q6PGN3 | Dclk2 | 1.80 | 1.1E-02 | |||
| P62631 | Eef1a2 | 1.51 | 2.5E-02 | [43] | ||
| Q91WK2 | Eif3h | 1.20 | 1.5E-02 | |||
| Q99LI8 | Hgs | 1.07 | 2.7E-02 | [44] | ||
| A2CG49 | Kalrn | 2.71 | 4.2E-02 | |||
| Q63844 | Mapk3 | 2.32 | 4.9E-02 | |||
| Q8R001 | Mapre2 | 3.22 | 3.8E-02 | |||
| Q80TL0 | Ppm1e | 2.02 | 5.9E-04 | |||
| Q8BG02 | Ppp2r2c | 1.75 | 4.3E-03 | |||
| P62983 | Rps27a (Ubiquitin) | 2.80 | 1.0E-04 | |||
| Q9D883 | U2af1 | 1.03 | 4.2E-02 |
1The proteins, denoted by Uniprot IDs and gene names, are significantly more abundant in the Q20 and/or Q140 samples with respect to the negative control. These proteins have log2 fold changes higher than 1 (at least two times more abundant) and a p-value less than 0.05 from the t-test.
Next, we performed a functional analysis on putative interactors of fl wtHtt and mHtt (Fig. 3). Several interactors of wtHtt have been linked with each other through previous studies, as shown by the network created using the STRING database (Fig. 3A). In this network, Syt1, Syt2, Snap47 and Ykt6 proteins formed a cluster, and these proteins are functional in vesicle fusion and exocytosis (Fig. 3B). Additionally, several of the putative interactors of mHtt are connected with each other based on the information in the STRING database (Fig. 3C). A functional analysis
Fig. 3
Functional analysis of proteins identified as wtHtt and mHtt interactors. A) Network created by the STRING database of proteins identified as interactors of wtHtt. B) Cluster of functionally related proteins identified by MCODE in the group of proteins that were identified as interactors of wtHtt. C) Network created by the STRING database from proteins identified as interactors of mHtt. D) Clusters of functionally related proteins identified by MCODE in the group of proteins that were identified as interactors of mHtt.
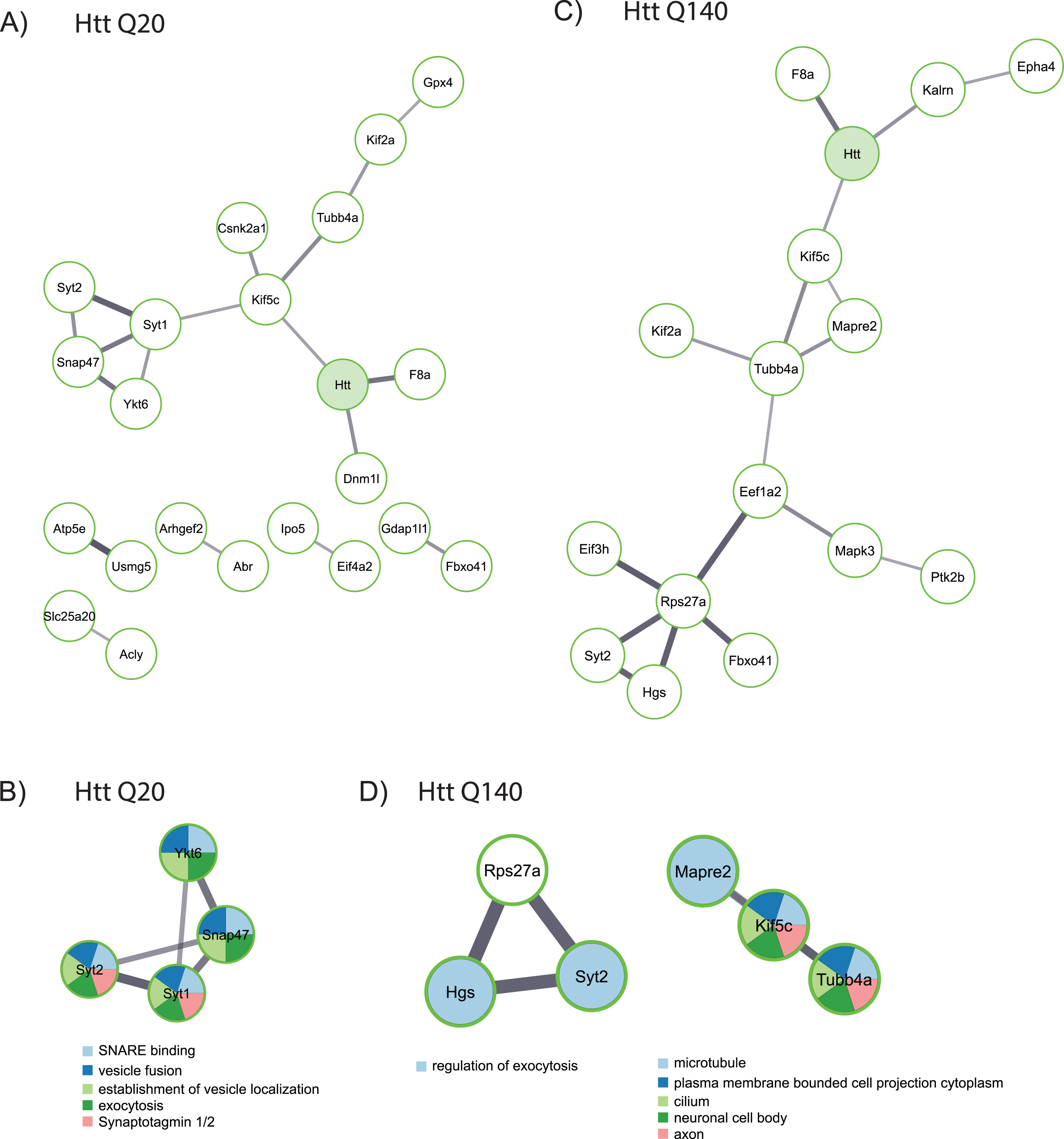
using MCODE shows that some of these proteins function in the regulation of exocytosis, while others play a role at the microtubule network and in the neuronal cell body (Fig. 3D).
DISCUSSION
WtHtt and mHtt have various common interaction partners in anterograde vesicle transport, but different interaction partners in processes like vesicle docking and exocytosis
In this study, we identified putative interactors of fl wtHtt and mHtt in two-month-old mice cortex brain regions. Table 1 lists all proteins that were significantly enriched in IPs for Flag-tagged fl Htt as compared to the non-Flag-tagged negative control. Some proteins were significantly enriched in both IPs for wtHtt and mHtt. This includes obviously the bait protein Htt, but also microtubule associated proteins, such as kinesins Kif5c and Kif2a. Kinesins are ATP-driven motor proteins that move along microtubules, thereby supporting several cellular functions including the transport of cargo, for instance in axonal transport. Neurons are particularly dependent on a functional transport system for organelles and molecules due to the long distance of axons. Most kinesins move towards the plus end of microtubules, which, in most cells, results in the transport of cargo from the center of the cell towards the periphery (anterograde transport). Cargo of kinesins include membrane bound organelles such as mitochondria, synaptic vesicles and synaptic vesicle precursors [21]. The putative interactor of both wtHtt and mHtt, kinesin heavy chain isoform 5C (Kif5C) is localized in the cytoplasm but is also abundant in the distal regions of dendrites and is involved in synaptic transmission. Additionally, the putative interactor of wtHtt and mHtt, kinesin-like protein Kif2a may regulate microtubule dynamics during axonal growth [22]. Tubb4a was also a common interactor of both wtHtt and mHtt. Tubb4a is the major subunit of microtubules [23]. It is well-known that Htt plays an important role in axonal transport [24] and our data shows that both wtHtt and mHtt interact with components of the anterograde transport system including kinesins and microtubules. However, we observed that Trim3, which is an E3 ubiquitin ligase localized at intracellular and Golgi-derived vesicles and can modulate the motor function of Kif21b [25], was interacting exclusively with wtHtt, but not with mHtt. Fast axonal transport is affected in polyglutamine diseases [26], and it has been suggested that polyQ proteins titrate motor proteins from other cargoes [24]. Our data shows that both wtHtt and mHtt can interact with proteins that play a role in membrane interactions and fusion during vesicle transport, for instance, Synaptotagmin 2 (Syt2). Synaptotagmins are transmembrane proteins involved in the regulation of membrane trafficking [27], and Syt2 was identified previously as Htt interactor in mice brain tissue [28, 29]. Various interacting proteins involved in membrane interactions and fusion were, however, unique for either wtHtt or mHtt. For instance, synaptotagmin 1 (Syt1), which may have a regulatory role in membrane interactions during synaptic vesicle trafficking in the active synapse zone was found to interact with wtHtt only. Interestingly, changes in expression levels of synaptotagmins have been linked to a variety of neurodegenerative diseases [27]. Additionally, Ykt6, which interacted with wtHtt in our screen, is part of the SNARE complex and mediates vesicle docking and fusion to a specific acceptor cellular compartment, for instance the endoplasmic reticulum or Golgi [30]. Finally, wtHtt interacted with Snap47, or synaptosomal-associated 47kDa protein, a protein that regulates brain-derived neurotrophic factor (BDNF) [31] and may also play a role in intracellular membrane fusion.
Unique interactors of mHtt include Hgs, or Hepatocyte growth factor-regulated tyrosine kinase substrate, which is a key component of the ESCRT-0 complex and recognizes ubiquitinated cargo and initiates the recruitment of endosomal sorting complexes. The ESCRT-0 complex allows for a specific membrane remodeling mode that results in the bending/budding of the membrane away from the cytoplasm [32]. Similarly, ubiquitin/Rps27 was a unique interactor of mHtt in our screen. We and others previously showed that mHtt is ubiquitinated more at its N-terminus compared to wtHtt [33, 34]. It is also possible that mHtt interacts with more ubiquitinated proteins. Concluding, our data suggests that both wtHtt and mHtt interact with kinesins and microtubules, which are important components of the anterograde transport system. In contrast, the proteins that we identified with a role in the delivery of cargo are interacting differently with wtHtt and mHtt, which might suggest that this process is regulated differently in HD mice as compared to wild-type mice. More research is required to confirm and investigate the role of the identified wtHtt and mHtt interaction partners in anterograde transport and cargo delivery processes.
WtHtt and mHtt interact with Hap40 which plays a role in endocytosis
Endocytosis is the process of cellular uptake of fluid and material from the extracellular space by enclosing it with a small portion of the plasma membrane and subsequent invagination and pinching off an endocytic vesicle into the interior of the cell. These vesicles are called early endosomes and they can mature to late endosomes which in turn can deliver their ingested particles to lysosomes or the Golgi complex. Early endosomes can be transported via actin for short distances, and via bidirectional transport along microtubules for longer distances [35]. The relatively most abundant interactor of both wtHtt and mHtt in our screen is F8a1, also known as Huntingtin-associated protein 40 (Hap40) (Table 1 and Fig. 2B). Hap40 is a known interactor of Htt and regulates the recruitment of Htt onto early endosomes. Htt and Hap40 together with Rab5 play an important role in the switching between actin and microtubule filaments for early endosome motility. It has been found that the microtubule-based transport of early endosomes is affected during HD pathology [36].
Unique interactions of wtHtt with proteins involved in energy metabolism and mitochondria
Several proteins that we identified as putative Htt interactors were unique to wtHtt and play a role in mitochondria, such as Dnm1l, an important mediator of mitochondrial and peroxisomal fission. Additionally, several wtHtt interactors are functional in mitochondria-mediated energy metabolism. For instance, Acly, or ATP citrate lyase, is an enzyme that plays a role in fatty acid biosynthesis [37]. The wtHtt interactor Slc25a20 is a mitochondrial carnitine/acylcarnitine translocase. It mediates the transport of acylcarnitines from the cytosol to the mitochondrial matrix to be oxidized by the mitochondrial fatty acid-oxidation pathway [38]. ATP5e, a component of the ATP synthase complex, or Complex V of the mitochondrial oxidative phosphorylation complex is also a putative interactor of wtHtt. Another putative wtHtt interactor, Usmg5 is a subunit of ATP synthase. There are links between ATP synthase and HD as overexpression of the ATP synthase α-subunit has been shown to reduce HD aggregation and toxicity [39].
Unique interactions of mHtt with proteins involved in translation
Several proteins that we identified as Htt interactors were unique to mHtt, including proteins with a role in the regulation of protein biosynthesis. For instance, serine/threonine kinase Mapk3, which is an essential component of the MAP kinase signal transduction pathway. This pathway can regulate many cellular processes, including translation [40]. Eif3h, or eukaryotic translation initiation factor 3 subunit H, was also identified as a mHtt interactor. Eif3h is a component of the eukaryotic translation initiation factor 3 (eIF-3) complex, which is needed during several steps in the initiation of protein synthesis [41]. Another unique putative interactor of mHtt in our screen is Eef1a2, or eukaryotic elongation factor 1 alpha 2. This protein binds to aminoacyl-tRNA to transport to it the A-site of ribosomes during protein synthesis [42]. Taken together, mHtt interacted with several proteins involved in protein translation while such interactions were not observed for wtHtt, which suggests that protein translation is affected by mHtt expression.
Several putative Htt interactors have been linked to HD and other neurodegenerative diseases
Additionally, we identified proteins with a wide variety of cellular functions as putative interactors of wtHtt and/or mHtt in our screen and some of these have already been linked to HD or other neurodegenerative diseases. Our list of putative Htt interactors showed overlaps with other IP- and mass spectrometry-based studies in HD models [43–45] (Table 1). Several of our interactors are also functionally linked to HD or other neurodegenerative diseases. For instance, Ptk2b/Pyk2 was identified as a putative interactor of both wtHtt and mHtt in our screen. Pyk2 is a non-receptor calcium-dependent protein-tyrosine kinase that is highly expressed in the hippocampus and modulates excitatory synapses. Reduced expression levels of Pyk2 were found in the hippocampus of HD mouse model R6/2 and in HD patients, which might contribute to cognitive impairments observed in HD [46]. Another putative interactor of both wtHtt and mHtt is a protein involved in the regulation of autophagy, Pip4k2c. It has been shown that the inhibition of Pip4kγ decreases the pathological effects of the mutant huntingtin [47]. Furthermore, tyrosine kinase ephrin receptor A4 (EphA4) that we identified as an interactor of wtHtt and mHtt, is known to mediate dendritic spine remodeling and retraction. This protein has been linked to Alzheimer’s disease (AD), as it functions as a receptor for amyloid beta oligomers which in turn leads to dendritic spine elimination [48]. The putative interactor of both wtHtt and mHtt, Fbxo41, is a neuron-specific F-box protein (a potential E3 ligase) that stimulates neuronal migration. Fbxo41 knock-out in mice displays a phenotype similar to ataxia with neuronal migration defects and degeneration of the cerebellum [49]. Another putative interactor of mHtt was Ahsa1. Ahsa1 is a co-chaperone of Hsp90aa1 and activates its ATPase activity which leads to an increase of its chaperone activity. Hsp90 and its co-chaperones are major regulators of protein folding and are involved in various cellular processes related to neurodegenerative diseases such as HD, AD, Parkinson’s disease, and prionopathies [50].
Future perspectives
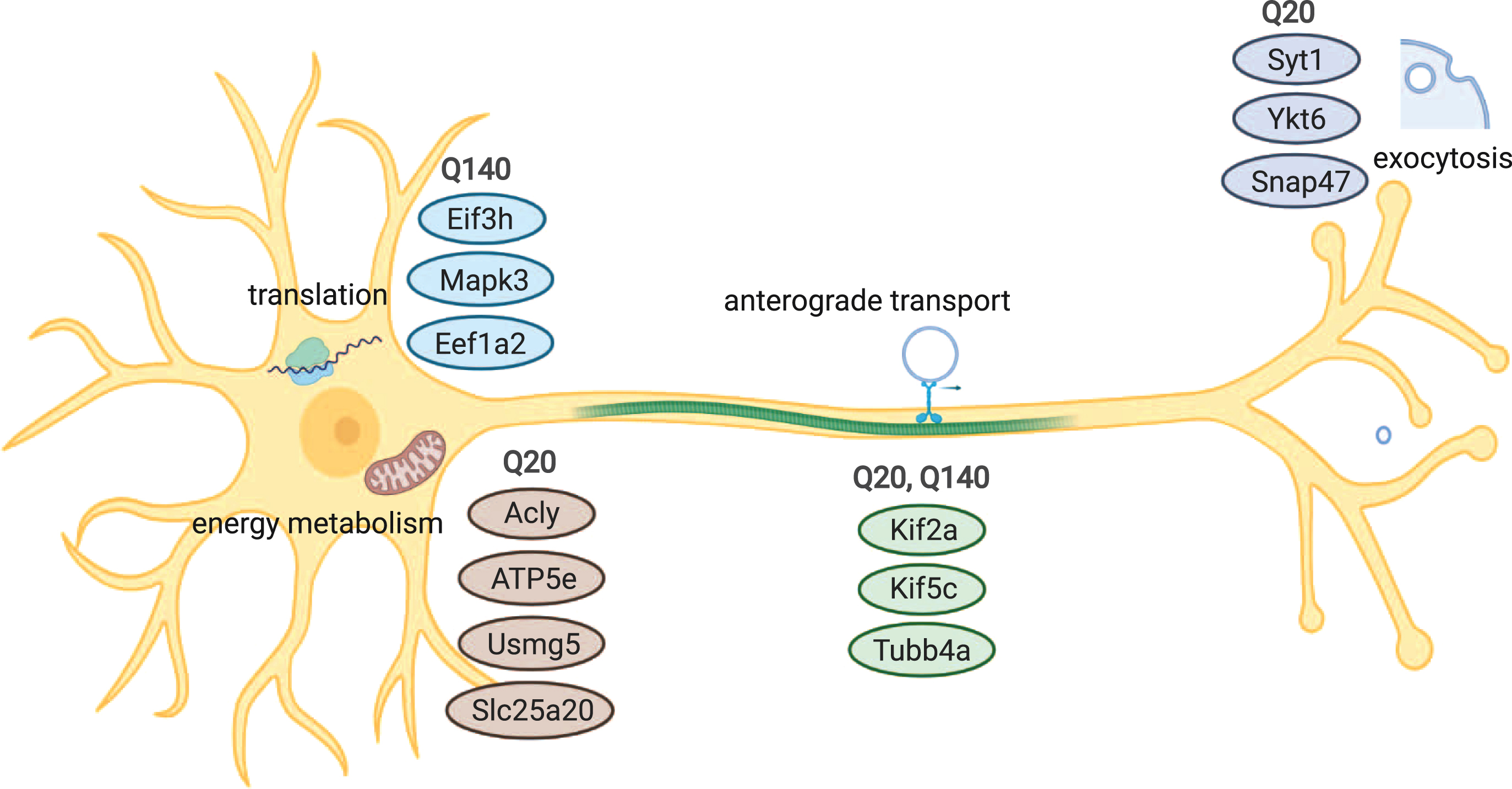
In this work we reported both shared and unique wtHtt and mHtt interactors, which are involved in various cellular processes, including exocytosis, transport of vesicles, translation and metabolism (Fig. 4). Further research could validate these interactions using different methods, identify binding sites and investigate the role of wtHtt and mHtt in more detail in the mentioned cellular processes. These findings will contribute to the understanding of the roles of wtHtt and mHtt in healthy conditions and HD pathology.
Fig. 4
Overview of cellular processes and components that are associated with wtHtt and/or mHtt interaction. Common interaction partners of wtHtt and mHtt play roles in anterograde transport, while proteins involved in exocytosis and vesicle docking differed between both forms of Htt. A group of interactors of wtHtt is involved in energy metabolism and mitochondria, while a group of interactors of mHtt is involved in protein translation.
ACKNOWLEDGMENTS
Figure 1 and Fig. 4 were created with BioRender.com. Figure 2C was created with VIB-UGENT Center for Plant Systems Biology Venn diagram tool on http://bioinformatics.psb.ugent.be/webtools/Venn/
This work was carried out on the Dutch national e-infrastructure with the support of SURF Cooperative. This work was funded by CHDI and Campagneteam Huntington.
CONFLICT OF INTEREST
The authors have no conflict of interest to report.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [51] with the dataset identifier PXD024254 and DOI 10.6019/PXD024254.
SUPPLEMENTARY MATERIAL
[1] The supplementary material is available in the electronic version of this article: https://dx.doi.org/10.3233/JHD-210476.
The supplementary tables have been uploaded to GitHub: https://github.com/ReitsGroup/FlagIP_JHD
REFERENCES
[1] | MacDonald ME , Ambrose CM , Duyao MP , Myers RH , Lin C , Srinidhi L , et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. (1993) ;72: (6):971–83. doi: 10.1016/0092-8674(93)90585-E |
[2] | Andrew SE , Paul Goldberg Y , Kremer B , Telenius H , Theilmann J , Adam S , et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington’s disease. Nat Genet. (1993) ;4: (4):398–403. doi: 10.1038/ng0893-398 |
[3] | Ross CA . When more is less: Pathogenesis of glutamine repeat neurodegenerative diseases. Neuron. (1995) ;15: (3):493–6. doi: 10.1016/0896-6273(95)90138-8 |
[4] | De La Monte SM , Vonsattel JP , Richardson EP . Morphometric demonstration of atrophic changes in the ceRebral cortex, white matter, and neostriatum in huntington’s disease. J Neuropathol Exp Neurol. (1988) ;47: (5):516–25. doi: 10.1097/00005072-198809000-00003 |
[5] | Li W , Serpell LC , Carter WJ , Rubinsztein DC , Huntington JA . Expression and characterization of full-length human huntingtin, an elongated HEAT repeat protein. J Biol Chem. (2006) ;281: (23):6–22. doi: 10.1074/jbc.M511007200 |
[6] | Brandstaetter H , Kruppa AJ , Buss F . Huntingtin is required for ER-to-Golgi transport and for secretory vesicle fusion at the plasma membrane. DMM Dis Model Mech. (2014) ;7: (12):1335–40. doi: 10.1242/dmm.017368 |
[7] | Orr AL , Li S , Wang CE , Li H , Wang J , Rong J , et al. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. (2008) ;28: (11):2783–92. doi: 10.1523/JNEUROSCI.0106-08.2008 |
[8] | Li JY , Plomann M , Brundin P . Huntington’s disease: A synaptopathy? Trends Mol Med. (2003) ;9: (10):414–20. doi: 10.1016/j.molmed.2003.08.006 |
[9] | Harjes P , Wanker EE . The hunt for huntingtin function: Interaction partners tell many different stories. Trends Biochem Sci. (2003) ;28: (8):425–33. doi: 10.1016/S0968-0004(03)00168-3 |
[10] | Sutherland BW , Toews J , Kast J . Utility of formaldehyde cross-linking and mass spectrometry in the study of protein-protein interactions. J Mass Spectrom. (2008) ;43: (43):699–715. doi: 10.1002/jms.1415 |
[11] | Zheng S , Ghitani N , Blackburn JS , Liu JP , Zeitlin SO . A series of N-terminal epitope tagged Hdh knock-in alleles expressing normal and mutant huntingtin: Their application to understanding the effect of increasing the length of normal huntingtins polyglutamine stretch onCAG140 mouse model pathogenesis. Mol Brain. (2012) ;5: (1):1. doi: 10.1186/1756-6606-5-28 |
[12] | Menalled LB , Sison JD , Dragatsis I , Zeitlin S , Chesselet MF . Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J Comp Neurol. (2003) ;465: (1):11–26. doi: 10.1002/cne.10776 |
[13] | Chalkley GE , Verrijzer CP . Immuno-depletion and purification strategies to study chromatin-remodeling factors in vitro. Methods Enzymol. (2004) ;377: (148):421–42. doi: 10.1016/S0076-6879(03)77028-1 |
[14] | Sap KA , Bezstarosti K , Dekkers DHW , van den Hout M , van Ijcken W , Rijkers E , et al. Global quantitative proteomics reveals novel factors in the ecdysone signaling pathway in Drosophila melanogaster. Proteomics. (2015) ;15: (4):725–38. doi: 10.1002/pmic.201400308 |
[15] | Tyanova S , Temu T , Cox J . The MaxQuant computational platform for mass spectrometry –based shotgun proteomics. Nat Protoc. (2016) ;11: (12):2301–19. doi: 10.1038/nprot.2016.136 |
[16] | Eden E , Navon R , Steinfeld I , Lipson D , Yakhini Z . GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. (2009) ;10: (48). doi: 10.1186/1471-2105-10-48 |
[17] | Supek F , Bošnjak M , Škunca N , Šmuc T . Revigo summarizes and visualizes long lists of gene ontology terms. PLoS One. (2011) ;6: (7):21800. doi: 10.1371/journal.pone.0021800 |
[18] | Doncheva NT , Morris JH , Gorodkin J , Jensen LJ . Cytoscape StringApp: Network analysis and visualization of proteomics data. J Proteome Res. (2019) ;18: (2):623–32. doi: 10.1021/acs.jproteome.8b00702 |
[19] | Shannon P , Markiel A , Owen Ozier 2 , Baliga NS , Wang JT , Ramage D , et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) ;(13):2498–504. doi: 10.1101/gr.1239303.metabolite |
[20] | Bader GD , Hogue CW . An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. (2003) ;4: :2. doi: 10.1186/1471-2105-4-2 |
[21] | Franker MAM , Hoogenraad CC . Microtubule-based transport -basic mechanisms, traffic rules and role in neurological pathogenesis. J Cell Sci. (2013) ;126: (11):2319–29. doi: 10.1242/jcs.115030 |
[22] | Niwa S . Kinesin superfamily proteins and the regulation of microtubule dynamics in morphogenesis. Anat Sci Int. (2015) ;90: (1). doi: 10.1007/s12565-014-0259-5 |
[23] | Duncan ID , Bugiani M , Radcliff AB , Moran JJ , Lopez-Anido C , Duong P , et al. A mutation in the Tubb4a gene leads to microtubule accumulation with hypomyelination and demyelination. Ann Neurol. (2017) ;81: (5):690–702. doi: 10.1002/ana.24930 |
[24] | Gunawardena S , Her LS , Brusch RG , Laymon RA , Niesman IR , Gordesky-Gold B , et al. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. (2003) ;40: (1):25–40. doi: 10.1016/S0896-6273(03)00594-4 |
[25] | Labonté D , Thies E , Pechmann Y , Groffen AJ , Verhage M , Smit AB , et al. TRIM3 regulates the motility of the kinesin motor protein KIF21B. PLoS One. (2013) ;8: (9):1–13. doi: 10.1371/journal.pone.0075603 |
[26] | Morfini G , Pigino G , Brady ST . Polyglutamine expansion diseases: Failing to deliver. Trends Mol Med. (2005) ;11: (2):64–70. doi: 10.1016/j.molmed.2004.12.002 |
[27] | Glavan G , Schliebs R , Živin M . Synaptotagmins in neurodegeneration. Anat Rec. (2009) ;292: (12):1849–62. doi: 10.1002/ar.21026 |
[28] | Culver BP , Savas JN , Park SK , Choi JH , Zheng S , Zeitlin SO , et al. Proteomic analysis of wild-type and mutant huntingtin-associated proteins in mouse brains identifies unique interactions and involvement in protein synthesis. J Biol Chem. (2012) ;287: (26):9–614. doi: 10.1074/jbc.M112.359307 |
[29] | Kaltenbach LS , Romero E , Becklin RR , Chettier R , Bell R , Phansalkar A , et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. (2007) ;3: (5):689–708. doi: 10.1371/journal.pgen.0030082 |
[30] | Nichols BJ , Pelham HRB . SNAREs and membrane fusion in the Golgi apparatus. Biochim Biophys Acta. (1998) ;1404: (1-2):9–31. doi: 10.1016/S0167-4889(98)00044-5 |
[31] | Yu C , Li CH , Chen S , Yoo H , Qin X , Park H . Decreased BDNF release in cortical neurons of a knock-in mouse model of Huntington’s disease. Sci Rep. (2018) ;8: (1):1–11. doi: 10.1038/s41598-018-34883-w |
[32] | Schuh AL , Audhya A . The ESCRT machinery: From the plasma membrane to endosomes and back again. Crit Rev Biochem Mol Biol. May. (2014) ;49: (3):242–61.. doi: 10.3109/10409238.2014.881777 |
[33] | Sap KA , Guler AT , Bezstarosti K , Bury AE , Juenemann K , Demmers JAA , et al. Global proteome and ubiquitinome changes in the soluble and insoluble fractions of Q175 huntington mice brains. Mol Cell Proteomics. (2019) ;18: (9):1705–20. doi: 10.1074/mcp.RA119.001486 |
[34] | Hakim-Eshed V , Boulos A , Cohen-Rosenzweig C , Yu-Taeger L , Ziv T , Kwon YT , et al. Site-specific ubiquitination of pathogenic huntingtin attenuates its deleterious effects. Proc Natl Acad Sci U S A. (2020) ;117: (31):18661–9. doi: 10.1073/pnas.2007667117 |
[35] | Granger E , McNee G , Allan V , Woodman P . The role of the cytoskeleton and molecular motors in endosomal dynamics. Semin Cell Dev Biol. (2014) ;31: :20–9. doi: 10.1016/j.semcdb.2014.04.011 |
[36] | Pal A , Severin F , Lommer B , Shevchenko A , Zerial M . Huntingtin-HAP40 complex is a novel Rab5 effector that regulates early endosome motility and is up-regulated in Huntington’s disease. J Cell Biol. (2006) ;172: (4):605–18. doi: 10.1083/jcb.200509091 |
[37] | Rhee J , Solomon LA , DeKoter RP . A role for ATP citrate lyase in cell cycle regulation during myeloid differentiation. Blood Cells, Mol Dis. (2019) ;76: (March):82–90. doi: 10.1016/j.bcmd.2019.02.006 |
[38] | Longo N , Frigeni M , Pasquali M . Carnitine transport and fatty acid oxidation. Biochim Biophys Acta. (2016) ;1863: (10):2422–35. doi: 10.1016/j.bbamcr.2016.01.023 |
[39] | Wang HQ , Xu YX , Zhao XY , Zhao H , Yan J , Sun XB , et al. Overexpression of F0F1-ATP synthase α suppresses mutant huntingtin aggregation and toxicity in vitro . Biochem Biophys Res Commun. (2009) ;390: (4):1294–8. doi: 10.1016/j.bbrc.2009.10.139 |
[40] | Roux PP , Topisirovic I . Signaling pathways involved in the regulation of mRNA translation. Mol Cell Biol. (2018) ;38: (12):1–26. doi: 10.1128/mcb.00070-18 |
[41] | Jivotovskaya A V ., Valášek L , Hinnebusch AG , Nielsen KH . Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol Cell Biol. (2006) ;26: (4):1355–72. doi: 10.1128/mcb.26.4.1355-1372.2006 |
[42] | Jakobsson ME , Małecki J , Falnes P . Regulation of eukaryotic elongation factor 1 alpha (eEF1A) by dynamic lysine methylation. RNA Biol. (2018) ;15: (3):314–9. doi: 10.1080/15476286.2018.1440875 |
[43] | Shirasaki DI , Greiner ER , Al-Ramahi I , Gray M , Boontheung P , Geschwind DH , et al. Network organization of the huntingtin proteomic interactome in mammalian brain. Neuron. (2012) ;75: (1):41–57. doi: 10.1016/j.neuron.2012.05.024 |
[44] | Hosp F , Vossfeldt H , Heinig M , Vasiljevic D , Arumughan A , Wyler E , et al. Quantitative interaction proteomics of neurodegenerative disease proteins. Cell Rep. (2015) ;11: (7):1134–46. doi: 10.1016/j.celrep.2015.04.030 |
[45] | Ratovitski T , Chighladze E , Arbez N , Boronina T , Herbrich S , Cole RN , et al. Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis. Cell Cycle. (2012) ;11: (10):2006–21. doi: 10.4161/cc.20423 |
[46] | Giralt A , Brito V , Chevy Q , Simonnet C , Otsu Y , Cifuentes-Díaz C , et al. Pyk2 modulates hippocampal excitatory synapses and contributes to cognitive deficits in a Huntington’s disease model. Nat Commun. (2017) ;8: (1):15592. doi: 10.1038/ncomms15592 |
[47] | Al-Ramahi I , Giridharan SSP , Chen YC , Patnaik S , Safren N , Hasegawa J , et al. Inhibition of PIP4Kγ ameliorates the pathological effects of mutant huntingtin protein. Elife. (2017) ;6: :1–25. doi: 10.7554/eLife.29123 |
[48] | Vargas LM , Cerpa W , Muñoz FJ , Zanlungo S , Alvarez AR . Amyloid-β oligomers synaptotoxicity: The emerging role of EphA4/c-Abl signaling in Alzheimer’s disease. Biochim Biophys Acta Mol Basis Dis.-59. (2018) ;1864: (4):1148–59. doi: 10.1016/j.bbadis.2018.01.023 |
[49] | Mukherjee C , Holubowska A , Schwedhelm-Domeyer N , Mitkovski M , Lee SJ , Kannan M , et al. Loss of the neuron-specific F-box protein FBXO41 models an ataxia-like phenotype in mice with neuronal migration defects and degeneration in the cerebellum. J Neurosci. (2015) ;35: (23):8701–17. doi: 10.1523/JNEUROSCI.2133-14.2015 |
[50] | Bohush A , Bieganowski P , Filipek A . Hsp90 and its co-chaperones in neurodegenerative diseases. Int J Mol Sci. (2019) ;20: (20). doi: 10.3390/ijms20204976 |
[51] | Perez-Riverol Y , Csordas A , Bai J , Bernal-Llinares M , Hewapathirana S , Kundu DJ , et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. (2019) ;47: (D1):D442–50. doi: 10.1093/nar/gky1106 |
[52] | Kegel KB , Meloni AR , Yi Y , Kim YJ , Doyle E , Cuiffo BG , et al. Huntingtin is present in the nucleus, interacts with the transcriptional corepressor C-terminal binding protein, and represses transcription. J Biol Chem. (2002) ;277: (9):7466–76. doi: 10.1074/jbc.M103946200 |
[53] | Goehler H , Lalowski M , Stelzl U , Waelter S , Stroedicke M , Worm U , et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington’s disease. Mol Cell. (2004) ;15: (6):853–65. doi: 10.1016/j.molcel.2004.09.016 |



