Diagnosis of COVID-19 based on chest X-ray images using pre-trained deep convolutional neural networks
Abstract
The novel coronavirus (COVID-19) that emerged and transmitted from China (Wuhan City) had a staggering effect on public health and the world economy. The early diagnosis of COVID-19 has become more important for its treatment and for controlling its spread due to its highly transmissible nature. In addition, the restricted supply of test kits calls for an alternative system for diagnosis. Since radiological images of chest of patients with COVID-19 show abnormalities, it is possible to diagnose COVID-19 utilizing chest X-ray images. Therefore, by applying deep convolution neural network (CNN), we have presented a diagnosis of COVID-19 based on chest X-ray images in this paper. For the diagnosis of COVID-19, an exhaustive comparative performance analysis of 16 state-of-the-art models is presented. Moreover, each model is trained with three approaches: transfer learning, fine tuning and scratch learning. The experiments were conducted on the dataset that comprises of 127 images of COVID-19, 500 images of Pneumonia and 500 images of normal cases. We have performed the experiments in two scenarios: binary classification (COVID-19 vs. Normal) and multiclass classification (COVID-19 vs. Pneumonia vs. Normal). Further, we have applied cost-sensitive learning technique to handle the class imbalance issue. In this study, InceptionResNetV2 model with fine-tuning approach achieved highest classification accuracy of 99.20% in binary classification and Xception model achieved classification accuracy of 89.33% in multiclass classification among all considered models. To validate our approach, we have presented the performance of our model on three other datasets and achieved adequate classification accuracy. Hence, the promising results demonstrate that the fine-tuning of deep CNN models is an effective way for diagnosis of COVID-19 and therefore, it can be deployed in diagnostic centers to assist radiologist after its validation with more prominent datasets.
1.Introduction
The COVID-19 is the most recently discovered pandemic disease caused by the coronaviruses. The outbreak of this disease began in China (Wuhan city) in December, 2019, thereafter it transmitted in many countries globally [1]. Initially, it is presumed that COVID-19 was infected by bat to human [2]. Coronavirus constitute a large family of viruses that causes sickness to animals and mankind. The disease caused by this virus transmits primarily from one person to another through tiny droplets either through mouth or nose, that are oozed out when a person with COVID-19 starts coughing, sneezing or speaking. The bigger droplets zoom through the air after the sneezing, but smaller droplets that are exhaled, are glided. The coronavirus is transmitted through air and quickly infect people when inhaled, causing serious illness. Different forms of coronavirus are responsible for respiratory infections, and symptoms vary from common cold to various diseases such as Middle East Respiratory Syndrome (MERS) and Severe Acute Respiratory Syndrome (SARS) [3]. The dry cough, fever and tiredness are the some of the unique occurrences in COVID-19 patients. Some people also get infected with mild symptoms such as headache, nasal congestions, and sore throat [4]. On the other hand, because of serious respiratory issues, few are associated with relatively high ICU admittance and mortality.
In current situation, researchers have been using various clinical methods for diagnosis of COVID-19. Reverse Transcription Polymerase Chain Reaction (RT-PCR) which is used for the gene sequencing of respiratory and blood samples [5], can also be used to detect COVID-19. However, as a result of low sensitivity of RT-PCR, COVID-19 cannot be detected quickly in patients, resulting lack of treatment which may result in infecting a large number of population [5]. Therefore, it is suggested that the computed tomography (CT) and chest X-ray (CXR) are sensitive methods and preferred to detect COVID-19 over RT-PCR [6]. These are most common techniques used for diagnosis of Pneumonia, lungs inflammation etc. In this study, CXR images are preferred and used to diagnosis COVID-19 over CT scan because of following reasons: (i) CXR imaging machines are easily available in hospitals, (ii) CXR images are cheaper than CT scan and (iii) CXR has low ionizing radiation than CT scan. However, it calls for radiology experts and takes significant time, which is key constraints in situation like COVID-19 pandemic. Therefore, many testing kits have been developed. However, we need to rely on other possible angle for its diagnosis due to limited number of testing kits. Therefore, developing an automated and reliable system for diagnosis of COVID-19 is necessary to save the professional’s precious time.
As per literature, several image based methods have been presented for detection of COVID-19. Ozturk et al. [7], proposed DarkCovidNet based on CNN using X-ray images for binary (COVID vs. No-Findings) and multiclass (COVID vs. Pneumonia vs. No-Findings) classification and achieved an accuracy of 98.08% and 87.02% respectively. Hemdan et al. [8] explored seven pre-trained deep CNN models for detection of healthy status against COVID-19 utilizing chest X-rays and obtained that performance of VGG16 and DenseNet201 is better compared to rest. Narin et al. [3] explored three deep learning models utilizing chest X-rays and achieved detection accuracy of 98% using ResNet50. Apostolopoulos et al. [9] explored six deep learning models with transfer learning approach using X-ray images and achieved classification accuracy of 96.78% for binary classification and 94.72% for multiclass classification. Sethy and Behera [10] used extracted features form different CNN models to feed into Support Vector Machine (SVM) for classification. In this experiment, ResNet50 model with SVM classifier achieved 95.38% accuracy. In the same line, Wang and Wong [11] proposed a COVID-Net that obtained 92.40% accuracy in detecting three classes i.e. normal, pneumonia and COVID-19 images. Das et al. [12] explored Xception with FT and obtained a classification accuracy of 97.40% while Pathak et al. [13] proposed a deep transfer learning and obtained an accuarcy of 93.02%. Nayak et al. [14] explored Resnet-34 to classify Normal and COVID-19 and achieved a classification accuracy of 98.33%. Gilanie et al. [15] proposed a CNN model for the classification of Normal, Pneumonia and COVID-19 images and reported an accuracy of 96.68%. Oh et al. [16] achieved a classification accuracy of 91.90% for classification of COVID-19, Pneumia and Normal using a patch-based CNN approach with limited training data.
In this paper, we have presented exhaustive comparison of performance of 16 state-of-the-art deep CNN models for binary classification (COVID-19 vs. Normal) and multiclass classification (COVID-19 vs. Pneumonia vs. Normal). The models used in this paper are: (i) VGG16, (ii) VGG19, (iii) ResNet50, (iv) ResNet101, (v) ResNet152, (vi) ResNet50V2, (vii) ResNet101V2, (viii) ResNet152V2, (vix) InceptionV3, (x) InceptionResNetV2, (xi) Xception, (xii) MobileNet, (xiii) MobileNetV2, (xiv) DenseNet121 (xv) DenseNet169, (xvi) DenseNet201. Further, the performances of these 16 models have been compared in three different training strategy: transfer learning (TL), fine tuning (FT) and scratch learning (SL). Therefore, this paper’s contributions are as follows: (i) diagnosis of COVID-19 using chest X-ray images; (ii) diagnosis of COVID-19 in two scenarios: binary classification (COVID-19 vs. Normal) and multiclass classification (COVID-19 vs. Pneumonia vs. Normal); (iii) performance comparison of 16 state-of-the-art deep CNN models in diagnosis of COVID-19; (iv) comparative performance analysis of models in three training strategies: TL, FT and SL; (v) handled class imbalance issue using cost-sensitive learning technique; (vi) achieved classification accuracy of 99.20% in binary classification and 89.33% in multiclass classification using InceptionResNetV2 and Xception model respectively with fine-tuning approach; (vii) validation of these models on three other datasets obtained from different sources.
The remainder of this paper is arranged as follows. Section 2 explains methodology and the state-of-the-art deep CNN models used in this paper. Section 3 presents the experimental result and performance analysis and finally, Section 4 concludes our study.
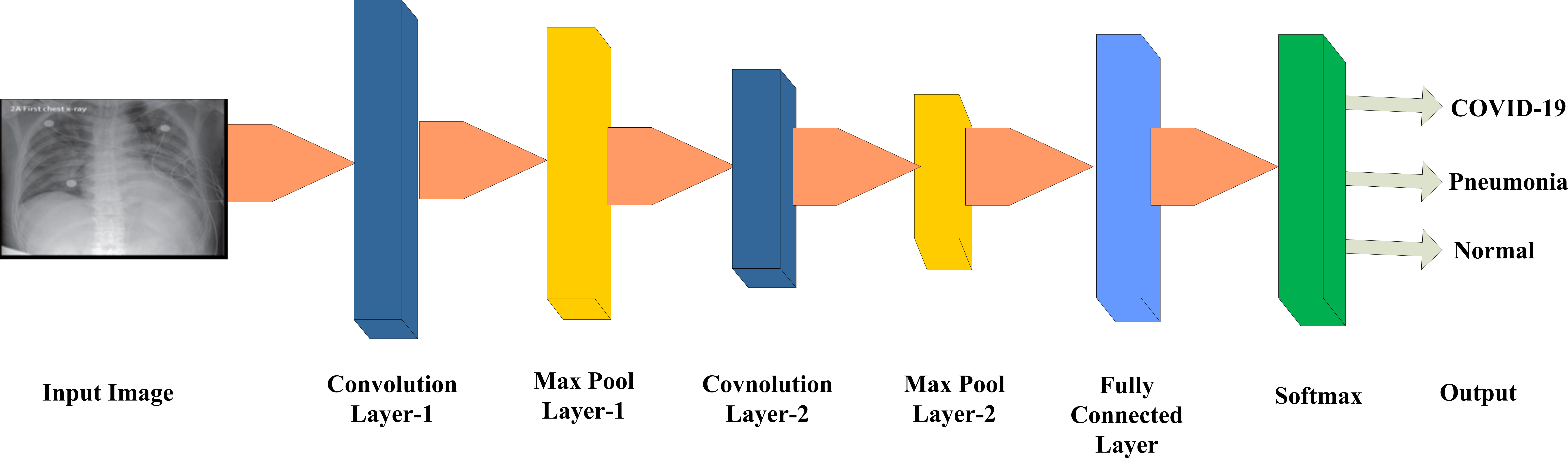
Figure 1.
General deep CNN architecture for diagnosis of COVID-19.
2.Methodology
A significant development of deep learning has given a new dimension in machine learning domain during the last decades. The convolutional neural network (CNN) has emerged as most powerful model among various deep learning models for visual recognition tasks [17, 18, 19]. Figure 1 depicts the general architecture of CNN for the diagnosis of COVID-19.
The primary components of CNN architecture are: (a) convolution layer, (b) pooling layer, and (c) fully connected layer. In the process of CNN, each iteration is called as an epoch. The trained model with deep CNN showed high discriminative power. The different components of it is described below.
a) Convolution layer
CNN’s main component is the convolution layer. It’s principle concept is to extract local features from the input image and generate feature maps [20]. It consists of learnable parameters called filters or kernels that is convolved with input image and expressed as Eq. (4).
(1)
where,
b) Pooling layer
In CNN architecture, the convolution layers are accompanied by pooling layers. The pooling layer are used for subsampling. These layers are often referred to as down sampling layers because the spatial size of the function map is reduced and thus the computational complexity is minimized. Max pooling [21] is the most common pooling technique and it is expressed in Eq. (2).
(2)
where,
c) Fully connected layer
After many convolution and pooling layers in CNN architecture, there is fully connected layer. The fully connected layer has neurons that are having connections to all activations of previous layer. The activations in this layer are computed in the form of affine transformations that involves matrix multiplication and bias offset. There may be more than one fully connected layer in any model. The most common activation function in fully connected layer is Softmax. The Softmax function calculates the probability distribution of output classes and it is expressed as
(3)
Table 1
The details of four CXR datasets considered in this paper
| Dataset used | No. of COVID-19 images | No. of normal images | No. of pneumonia images | Total images |
|---|---|---|---|---|
| [7] | 125 | 500 | 500 | 1125 |
| [33] | 504 | 81 | 18 | 603 |
| [35] | 58 | 127 | – | 185 |
| [36] | 35 | 03 | 02 | 40 |
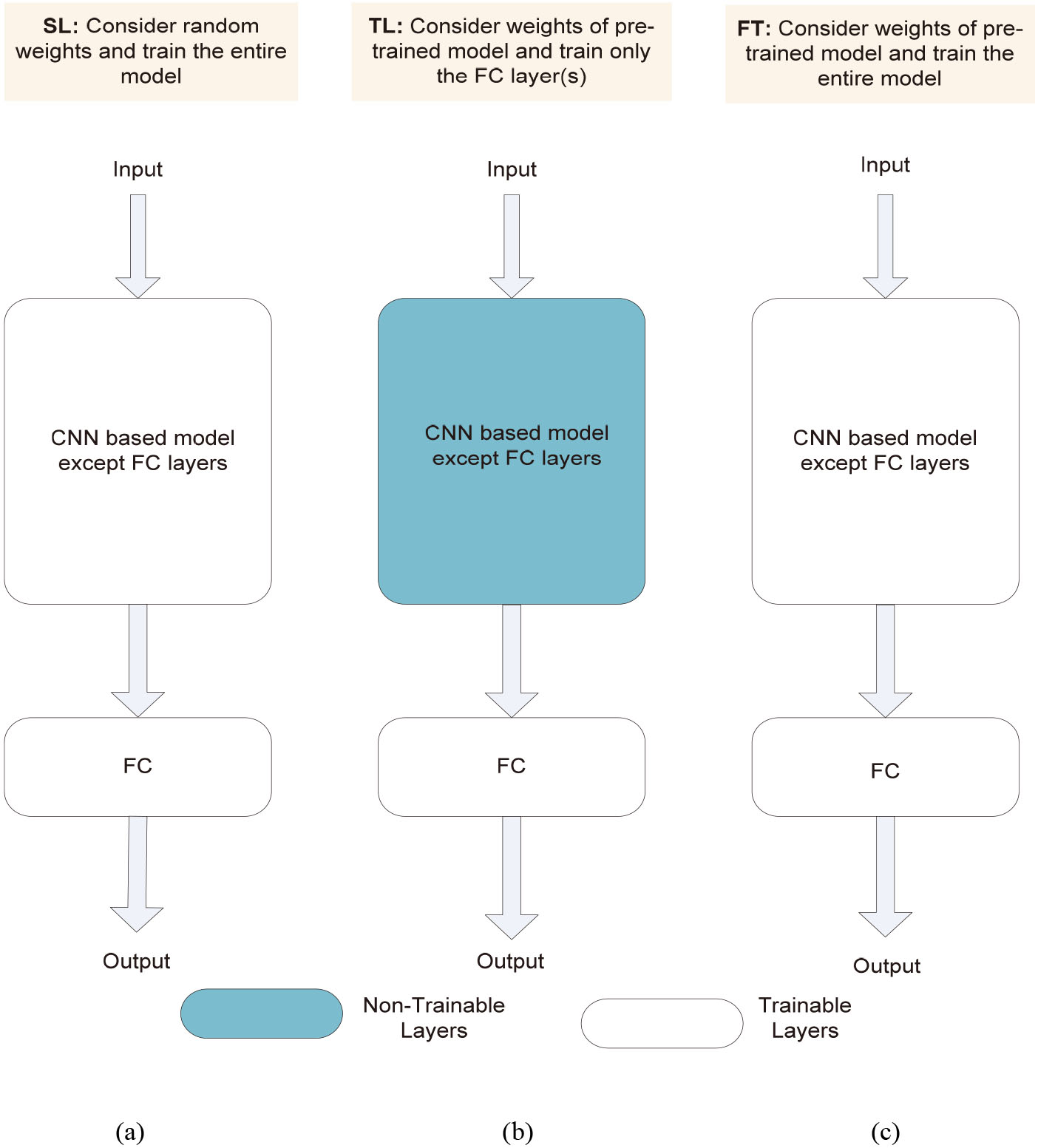
Figure 2.
General architecture of CNN model with three training approaches: (a) scratch learning; (b) transfer learning; (c) fine tuning.
where
All these layers are stacked to build a complete CNN architecture. In addition to these layers, the other layers such as batch normalization and dropout may be appended to reduce time complexity and avoid overfitting, respectively.
2.1Dataset description
In this paper, we have used four CXR image datasets obtained from different sources. The details of these datasets has been presented in Table 1. However, we have utilized CXR image dataset of [7] to train and explore the performance of 16 models. Figure 2 depicts the sample image of each class of this dataset [7]. The best performing model among 16 models were applied on three other datasets [22, 23, 24] for validation.
2.2Transfer learning, fine tuning and scratch learning
The simplest way of training any CNN model is scratch learning. Here, the weights of all the layers are randomly initialized and updated using backpropagation algorithm through several iterations until it attains the minimum loss. Figure 2a shows the general architecture of a CNN model with scratch learning. However, it increases the computational cost and powerful GPU requirement as the number of layer increases. Therefore, techniques such as transfer learning and fine tuning has been proposed to solve these issues.
Availability of large dataset is not always possible and training with small dataset may cause the problem of overfitting. A transfer learning approach [25] has been introduced as an solution to this issue. Transfer learning is a method in which pre-trained model information is applied to a large dataset to solve a similar problem in different datasets. Here, the weights of all the layers are frozen except the last few fully connected layers, i.e., only the weights of unfrozen layers take part in training. Hence, it reduces the computational complexity while producing adequate performance. Figure 2b shows the general architecture of CNN model with transfer learning. Another technique called fine tuning is a mid-way approach between scratch learning and transfer learning. Here, the weights of pre-trained model are used as initial weights and the weights of all the layers are updated using backpropagation algorithm while training. Figure 2c shows the general architecture of CNN model with fine tuning.
2.3Pre-trained deep CNN models
We have explored 16 pre-trained deep CNN models for diagnosis of COVID-19 and they are: (i) VGG16, (ii) VGG19, (iii) ResNet50, (iv) ResNet101, (v) ResNet152, (vi) ResNet50V2, (vii) ResNet101V2, (viii) ResNet152V2, (ix) InceptionV3, (x) InceptionResNetV2, (xi) Xception, (xii) MobileNet, (xiii) MobileNetV2, (xiv) DenseNet121, (xv) DenseNet169, (xvi) DenseNet201. All of the aforementioned models have been pre-trained on ImageNet dataset [26] that consists of 1.2 million images of 1000 classes. Moreover, we have presented the exhaustive comparative performance analysis of these models in three training approaches: transfer learning, fine tuning and scratch learning. A brief description of each model has been provided below.
Simonyan et al., [27] proposed VGG16 model. It is a sequential CNN using 3
Figure 3.
Architecture of state-of-the-art deep CNN models.
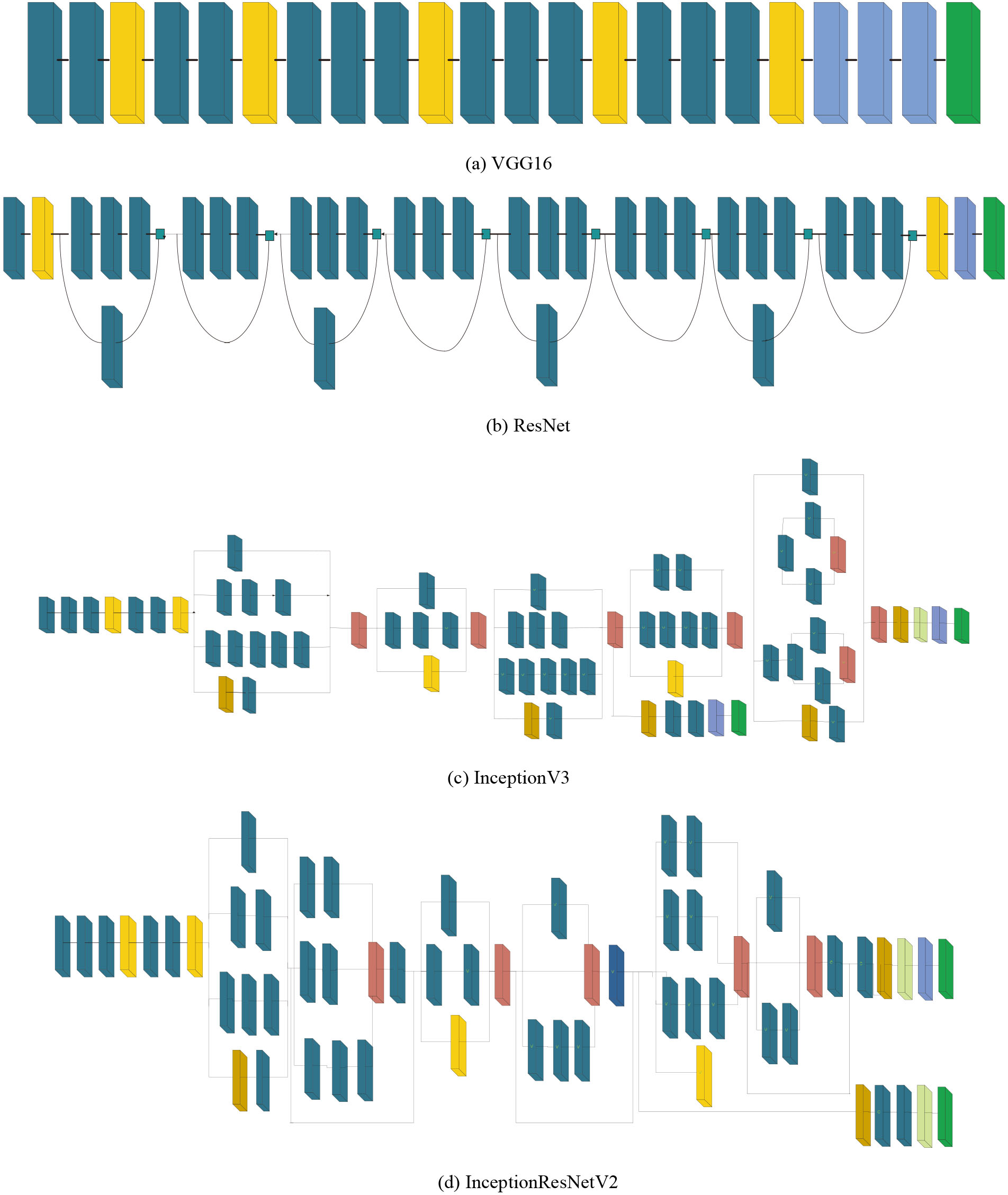
Figure 3.
Continued.
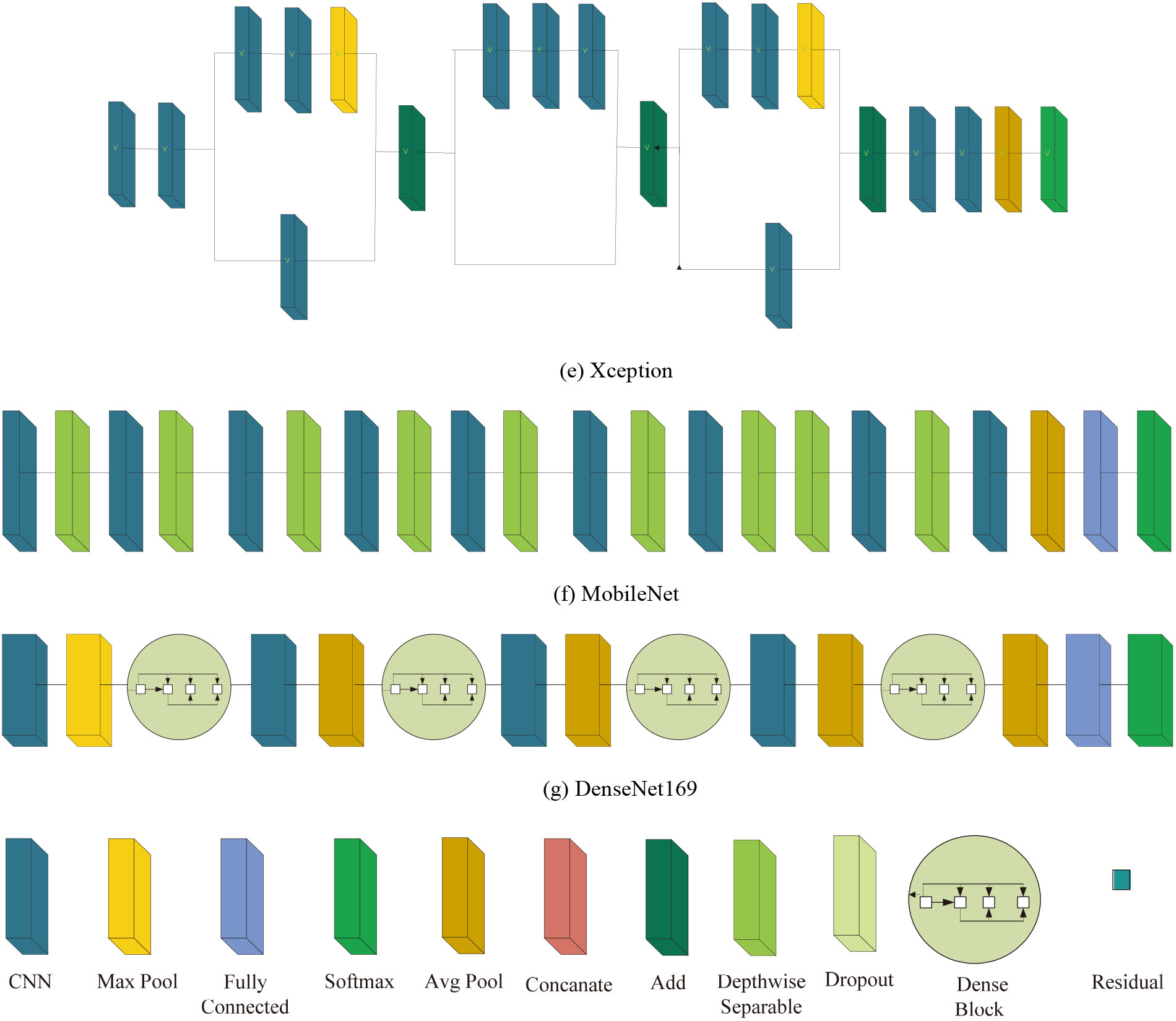
Figure 4.
Sample image of each class: (a) COVID-19, (b) Pneumonia and (c) Normal.
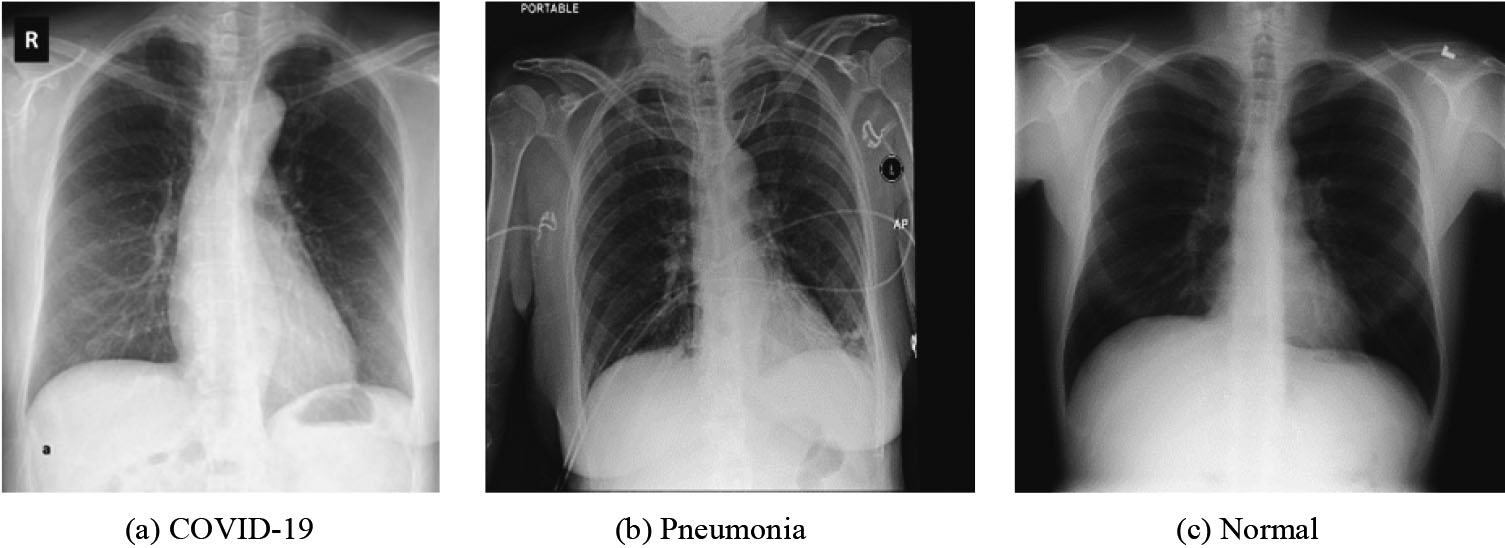
Table 2
Trainable parameters of deep CNN models
| Model | Binary classification | Multiclass classification | ||||
|---|---|---|---|---|---|---|
| TL | FT | SL | TL | FT | SL | |
| VGG16 | 8,194 | 134,268,738 | 134,268,738 | 12,291 | 134,272,835 | 134,272,835 |
| VGG19 | 8,194 | 139,578,434 | 139,578,434 | 12,291 | 139,582,531 | 139,582,531 |
| ResNet50 | 4,098 | 23,538,690 | 23,538,690 | 6,147 | 23,540,739 | 23,540,739 |
| ResNet101 | 4,098 | 42,556,930 | 42,556,930 | 6,147 | 42,558,979 | 42,558,979 |
| ResNet152 | 4,098 | 58,223,618 | 58,223,618 | 6,147 | 58,225,667 | 58,225,667 |
| ResNet50V2 | 4,098 | 23,523,458 | 23,523,458 | 6,147 | 23,525,507 | 23,525,507 |
| ResNet101V2 | 4,098 | 42,532,994 | 42,532,994 | 6,147 | 42,535,043 | 42,535,043 |
| ResNet152V2 | 4,098 | 58,192,002 | 58,192,002 | 6,147 | 58,194,051 | 58,194,051 |
| InceptionV3 | 4,098 | 21,772,450 | 21,772,450 | 6,147 | 21,774,499 | 21,774,499 |
| InceptionResNetV2 | 3,074 | 54,279,266 | 54,279,266 | 4,611 | 54,280,803 | 54,280,803 |
| Xception | 4,098 | 20,811,050 | 20,811,050 | 6,147 | 20,813,099 | 20,813,099 |
| MobileNet | 2,002 | 4,233,978 | 4,233,978 | 3,003 | 4,234,979 | 4,234,979 |
| MobileNetV2 | 2,562 | 2,226,434 | 2,226,434 | 3,843 | 2,227,715 | 2,227,715 |
| DenseNet121 | 2,050 | 6,955,906 | 6,955,906 | 3,075 | 6,956,931 | 6,956,931 |
| DenseNet169 | 3,330 | 12,487,810 | 12,487,810 | 4,995 | 12,489,475 | 12,489,475 |
| DenseNet201 | 3,842 | 18,096,770 | 18,096,770 | 5,763 | 18,098,691 | 18,098,691 |
2.4Class balancing
It can be observed from Table 1 that all four dataset has the class imbalance issue. For example, the number of COVID-19 images in the datasets of [7] are 125 whereas the other two classes (Pneumonia and Normal) have 500 images each. This results in bias to the performance of the model. Hence, we have used cost-sensitive learning [37] to handle the class imbalance problem which computes the class weights for each class as shown in Eq. (4) where, C represents number of classes and
(4)
3.Results and discussion
3.1Experimental design
The experimental setup to train all 16 deep CNN models has been described in this section. These are pre-trained models and already trained on a large dataset known as ImageNet. The ImageNet comprises of 1000 classes. Hence, these CNN models are having 1000 neurons in the FC layer to predict 1000 classes. However, we have evaluated for binary class (COVID-19 Vs. Normal) and multiclass (COVID-19 Vs. Pneumonia Vs. Normal) in this paper. Therefore, FC layer was replaced by two neurons in case of binary class and three neurons in case of multiclass. The size of the input image was reshaped as defined for each pre-trained CNN models. The size of the input image for InceptionV3, InceptionResNetV2 and Xception is 299
3.2Experimental results
The classification accuracy obtained using 16 models on test set has been shown in Tables 3 and 4 for binary and multiclass classification, respectively. Following observations have been made: (i) among three techniques (TL, FT, SL), SL has produced lowest accuracy in both scenario (binary and multiclass). The rationale behind it is that SL technique might require more number of epochs to effectively train the model. When comparing TL and FT, TL has performed better for few models and FT has produced better results for few other models. However, the difference in OA is very less in these two techniques; (ii) highest OA of 99.20% has been obtained using InceptionResNetV2 model with FT for binary classification (Table 3) and the highest OA of 89.33% has been obtained using Xception model with FT approach for multiclass classification (Table 4).
Table 3
Performance comparison of deep CNN models with TL, FT and SL approach for binary classification (COVID-19 vs. Normal)
| Deep CNN model | TL | FT | SL |
|---|---|---|---|
| Vgg16 | 97.12 | 90.56 | 80.48 |
| Vgg19 | 96.64 | 86.56 | 79.20 |
| ResNet50 | 83.52 | 94.08 | 91.84 |
| ResNet101 | 85.12 | 92.48 | 93.44 |
| ResNet152 | 78.24 | 93.12 | 90.88 |
| ResNet50V2 | 96.64 | 94.08 | 93.28 |
| ResNet101V2 | 97.60 | 96.80 | 91.84 |
| ResNet152V2 | 96.96 | 95.52 | 92.32 |
| InceptionV3 | 95.68 | 97.44 | 95.68 |
| InceptionResNetV2 | 97.12 |
99.20 | 94.40 |
| Xception | 97.60 | 97.60 | 98.40 |
| MobileNet | 95.36 | 97.92 | 93.44 |
| MobileNetV2 | 97.12 | 93.44 | 78.24 |
| DenseNet121 | 97.12 | 97.60 | 93.60 |
| DenseNet169 | 97.60 | 93.60 | 94.40 |
| DenseNet201 | 98.40 | 94.40 | 94.56 |
Table 4
Performance comparison of deep CNN models with TL, FT and SL approach for multiclass classification (COVID-19 vs. Pneumonia vs. Normal)
| Deep CNN model | TL | FT | SL |
|---|---|---|---|
| Vgg16 | 78.49 | 73.33 | 42.13 |
| Vgg19 | 78.76 | 57.42 | 45.24 |
| ResNet50 | 61.96 | 77.33 | 71.20 |
| ResNet101 | 60.36 | 82.40 | 68.80 |
| ResNet152 | 59.20 | 69.25 | 74.76 |
| ResNet50V2 | 80.18 | 83.11 | 73.16 |
| ResNet101V2 | 77.06 | 80.80 | 73.24 |
| ResNet152V2 | 80.89 | 81.78 | 69.24 |
| InceptionV3 | 78.49 | 86.22 | 77.16 |
| InceptionResNetV2 | 79.73 | 85.16 | 70.40 |
| Xception | 77.87 |
89.33 | 77.51 |
| MobileNet | 79.29 | 85.96 | 65.78 |
| MobileNetV2 | 81.42 | 67.47 | 44.53 |
| DenseNet121 | 80.89 | 82.84 | 74.31 |
| DenseNet169 | 81.07 | 84.18 | 68.09 |
| DenseNet201 | 80.89 | 84.27 | 77.87 |
Figure 5.
Confusion matrix: (a) Binary classification, (b) multiclass classification.
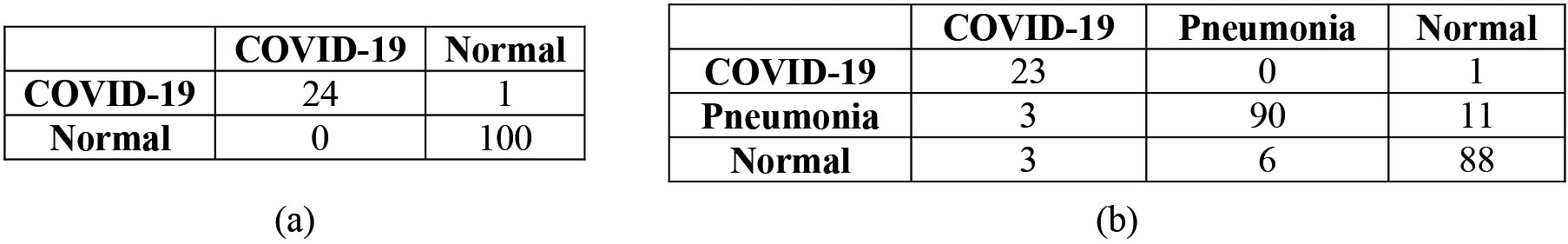
Figure 6.
ROC curve: (a) Binary classification, (b) multiclass classification.
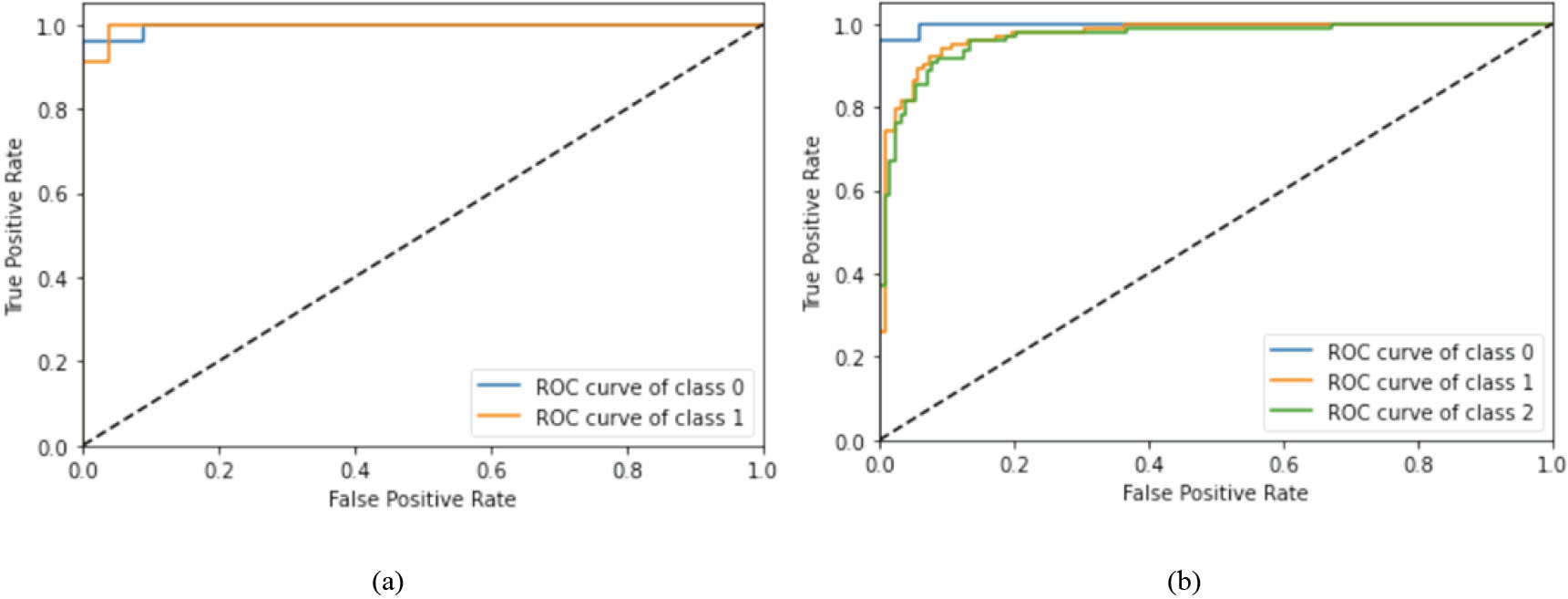
Table 5
Other performance parameters for binary and multiclass classification without class balancing
| Classes | Class accuracy (%) | Precision | Recall | F1-score |
|---|---|---|---|---|
| Binary classification | ||||
| COVID-19 | 96.00 | 1.00 | 0.96 | 0.98 |
| Normal | 100 | 0.99 | 1.00 | 1.00 |
| Multiclass classification | ||||
| COVID-19 | 95.83 | 0.79 | 0.96 | 0.87 |
| Pneumonia | 86.54 | 0.94 | 0.87 | 0.90 |
| Normal | 90.72 | 0.88 | 0.91 | 0.89 |
Table 6
Other performance parameters for binary and multiclass classification with class balancing
| Classes | Class accuracy (%) | Precision | Recall | F1-score |
|---|---|---|---|---|
| Binary classification | ||||
| COVID-19 | 97.00 | 0.97 | 0.97 | 0.97 |
| Normal | 99.00 | 0.99 | 0.99 | 0.99 |
| Multiclass classification | ||||
| COVID-19 | 100 | 0.96 | 1.00 | 0.98 |
| Pneumonia | 89.81 | 0.90 | 0.90 | 0.90 |
| Normal | 87.23 | 0.88 | 0.87 | 0.88 |
Table 7
Validation of our approach on other three CXR datasets with class balancing
| Data set used | No. of COVID-19 images | No. of normal images | No. of pneumonia images | Classification type | Model | Accuracy (%) |
|---|---|---|---|---|---|---|
| [22] | 504 | 81 | – | Binary | InceptionResNetV2 | 96.19 |
| 504 | 81 | 18 | Multiclass | Xception | 82.37 | |
| [23] | 58 | 127 | – | Binary | InceptionResNetV2 | 91.89 |
| [24] | 35 | 3 | – | Binary | InceptionResNetV2 | 91.67 |
| 35 | 3 | 2 | Multiclass | Xception | 91.68 |
Table 8
Benchmarking of our approach with existing approaches on COVID-19 diagnosis
| Authors | Modality | # classes with data size | Methodology | Classification accuracy (%) |
|---|---|---|---|---|
| Ozturk et al. [7] | CXR | 500 Normal 125 COVID-19 500 Pneumonia 500 Normal 125 COVID-19 | DarkCovidNet | 98.08 (Binary Class) 87.02 (Multiclass) |
| Narin et al. [3] | CXR | 50 COVID-19 50 Normal | ResNet50 with TL | 98.00 (Binary Class) |
| Apostolopoulos et al. [9] | CXR | 224 COVID-19 714 Bacterial and Viral Pneumonia 504 Normal | MobileNetV2 with TL | 96.78 (Binary Class) 94.72% (Multiclass) |
| Sethy et al. [10] | CXR | 133 COVID-19 133 Normal | ResNet50 with TL and SVM | 95.38 (Binary Class) |
| Hemdan et al. [8] | CXR | 25 COVID-19 25 Normal | COVIDX-Net | 90.00 (Binary Class) |
| Wang et al. [11] | CXR | 16,756 Images of Normal, Pneumonia and COVID-19 | COVID-Net | 92.40 (Multiclass) |
| Elasnaoui et al. [38] | CXR | 2780 Bacteria Pneumonia 1493 Coronavirus, 231 COVID-19 1583 Normal | InceptionResNetV2 with TL | 92.18 (Multiclass) |
| Das et al. [12] | CXR | 500 Pneumonia 500 Normal 125 COVID-19 | Extreme version of Inception | 97.40 (Multiclass) |
| Pathak et al. [13] | CXR | 419 COVID-19 439 Normal or Pneumonia infected | Deep Transfer Learning | 93.02 (Binary class) |
| Nayak et al. [14] | CXR | 203 Normal 203 COVID-19 | Resnet-34 | 98.33 (Binary Class) |
| Gilanie et al. [15] | CXR and CT | 7021 Normal and Pneumonia 1066 COVID-19 | CNN model | 96.68 (Binary class) |
| Oh et al. [16] | CXR | 5000 images (Normal, Pneumonia and COVID-19) | Patch-based CNN | 91.9 (Multiclass) |
| Our Approach | CXR | 500 Normal 125 COVID-19 500 Pneumonia 500 Normal 125 COVID-19 | InceptionResNetV2 with FT for Binary Class Xception with FT for multiclass | 99.20 (Binary Class) 89.33 (Multiclass) |
For a more detailed analysis, we have shown few other performance parameters using best performing models only, i.e., InceptionResNetV2 model for binary classification and Xception model for multiclass classification. Figure 5 depicts the confusion matrix for binary and multiclass classification where diagonal elements illustrate correct classification. Further, class accuracy, precision, recall and F1 score have been presented in Table 5 which shows our model is able to classify COVID-19 class with very high accuracy (96% in case of binary class and 95.83% in case of multiclass). Moreover, the receiving operating characteristic (ROC) curve has been shown in Fig. 6 where class 0, 1 and 2 represents COVID-19, Pneumonia and Normal class respectively. As all curves are closer to top-left corner, it demonstrates encouraging performance of the model. Further, we have applied cost-sensitive learning technique to handle the class imbalance issue and the results were shown in Table 6. It can be observed from Tables 5 and 6 that there is an improvement in COVID-19 classification accuracy after class balancing in case of binary classification (1.04% improvement) and multiclass (4.35% improvement) as well. Further, we have validated our approach on three other datasets obtained from different sources and the performance has been depicted in Table 7. From Table 7, it is evident that our approach is able to achieve adequate classification accuracy on other three datasets as well.
Lastly, to highlight the performance of our approach, a benchmarking of our approach with the existing approaches has been presented in Table 8. DarkCovidNet proposed by Ozturk et al., [7] for classification of COVID-19 obtained an accuracy of 98.08% and 87.02% for binary and multiclass respectively. Narin et al. [3] explored ResNet50 with TL and reported an accuracy of 98% while Sethy and Behera [10] explored ResNet50 with TL and SVM and achieved an accuracy of 94.72%. Apostolopoulos and Mpesiana [9] demonstared MobileNetV2 with TL and reported 96.78% accuracy for binary class and 94.72% accuracy for multiclass. COVIDX-Net proposed by Hemdan et al. [8], achieved an accuracy of 90% and COVID-Net proposed by Wang and Wond [11] obtained an accuracy of 92.40%. Elasnaoui and Chawki [38] explored InceptionResNetV2 with TL and reported 92.18% accuracy. Das et al. [12] explored Xception with FT and reported a classification accuracy of 97.40%. Pathak et al. [13] proposed a deep transfer learning (DTL) and reported an accuarcy of 93.02%. Nayak et al. [14] explored Resnet-34 and achieved a classification accuracy of 98.33%. Gilanie et al. [15] proposed a CNN model and reported an accuracy of 96.68%. Oh et al. [16] proposed a patch-based CNN and achieved a classification accuracy of 91.90% for classification of COVID-19, Pneumia and Normal class.
4.Conclusion
Due to the rapid growth in COVID-19 patients globally, automatic detection of COVID-19 patients is the need of the hour. A comprehensive comparative analysis of 16 deep CNN models to diagnose COVID-19 utilizing chest X-ray images has been presented in this paper. Further, the performances of these 16 models have been evaluated with three approaches namely TL, FT and SL in two scenarios: binary classification (COVID-19 vs. Normal) and multiclass classification (COVID-19 vs. Pneumonia vs. Normal). Our analysis concludes that the performance of deep CNN model is better with FT approach as compared to TL and SL for considered dataset. Among 16 models, InceptionResNetV2 has achieved highest classification accuracy of 99.20% in case of binary class and Xception model has obtained highest classification accuracy of 89.33% in case of multiclass. In addition, the class imbalance issue has been taken care using cost-sensitive learning technique and found improvement in COVID-19 classification accuracy. To substantiate the performance of our approach, experiments have been performed on other three datasets as well and observed that it achieved an adequate classification accuracy for all three datasets. In future, we intend to work on large datasets to validate our model. Subsequently, it may help radiologists to have a second opinion and prioritize their patients.
Conflict of interest
None.
References
[1] | Singh D, Kumar V, Yadav V, Kaur M. Deep Neural Network-Based Screening Model for COVID-19-Infected Patients Using Chest X-Ray Images. Int J Pattern Recognit Artif Intell. World Scientific. (2020) ; 2151004. |
[2] | Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. Elsevier. (2020) ; 395: (10223): 497–506. |
[3] | Narin A, Kaya C, Pamuk Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv Prepr arXiv200310849. (2020) . |
[4] | Singhal T. A review of coronavirus disease-2019 (COVID-19). Indian J Pediatr. Springer. (2020) ; 1–6. |
[5] | Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. Radiological Society of North America. (2020) ; 200642. |
[6] | Ng M-Y, Lee EYP, Yang J, Yang F, Li X, Wang H, et al. Imaging profile of the COVID-19 infection: Radiologic findings and literature review. Radiol Cardiothorac Imaging. Radiological Society of North America. (2020) ; 2: (1): e200034. |
[7] | Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. Elsevier. (2020) ; 103792. |
[8] | Hemdan EE-D, Shouman MA, Karar ME. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv Prepr arXiv200311055. (2020) . |
[9] | Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. Springer. (2020) . |
[10] | Sethy PK, Behera SK. Detection of coronavirus disease (covid-19) based on deep features. Preprints. (2020) ; 2020030300. |
[11] | Wang L, Wong A. COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from ChestX-Ray Images. arXivPrepr arXiv200309871. (2020) . |
[12] | Das NN, Kumar N, Kaur M, Kumar V, Singh D. Automated deep transfer learning-based approach for detection of COVID-19 infection in chest X-rays. IRBM. Elsevier. (2020) . |
[13] | Pathak Y, Shukla PK, Tiwari A, Stalin S, Singh S, Shukla PK. Deep Transfer Learning based Classification Model for COVID-19 Disease. IRBM. Elsevier. (2020) . |
[14] | Nayak SR, Nayak DR, Sinha U, Arora V, Pachori RB. Application of deep learning techniques for detection of COVID-19 cases using chestX-ray images: A comprehensive study. Biomed Signal Process Control. Elsevier. (2021) ; 64: : 102365. |
[15] | Gilanie G, Bajwa UI, Waraich MM, Asghar M, Kousar R, Kashif A, et al. Coronavirus (COVID-19) detection from chest radiology images using convolutional neural networks. Biomed Signal Process Control. Elsevier. (2021) ; 66: : 102490. |
[16] | Oh Y, Park S, Ye JC. Deep learning covid-19 features on CXR using limited training data sets. IEEE Transactions on Medical Imaging. (2020) ; 39: (8): 2688–2700. |
[17] | Jain N, Chauhan A, Tripathi P, Moosa SB, Aggarwal P, Oznacar B. Cell image analysis for malaria detection using deep convolutional network. Intell Decis Technol. IOS Press. (2020) ; 14: (1): 55–65. |
[18] | Shrivastava VK, Pradhan MK, Thakur MP. Application of Pre-Trained Deep Convolutional Neural Networks for Rice Plant Disease Classification. In: 2021 International Conference on Artificial Intelligence and Smart Systems (ICAIS). IEEE. (2021) ; pp. 1023–30. |
[19] | Shrivastava VK, Pradhan MK. Deep convolutional neural network based diagnosis of COVID-19 using x-ray images. Modelling and Analysis of Active Biopotential Signals in Healthcare. (2021) ; (2): 13–17. |
[20] | Wan J, Wang D, Hoi SCH, Wu P, Zhu J, Zhang Y, et al. Deep learning for content-based image retrieval: A comprehensive study. In: Proceedings of the 22nd ACM International Conference on Multimedia. (2014) ; pp. 157–66. |
[21] | Bera S, Shrivastava VK. Effect of pooling strategy on convolutional neural network for classification of hyperspectral remote sensing images. IET Image Process. IET. (2019) ; 14: (3): 480–6. |
[22] | Cohen JP, Morrison P, Dao L, Roth K, Duong TQ, Ghassemi M. COVID-19 Image Data Collection: Prospective Predictions Are the Future. arXiv Prepr arXiv200611988. (2020) . |
[23] | Chung A. Actualmed COVID-19 chest x-ray data initiative. https://github.com/agchung/Actualmed-COVID-chestxray-dataset. (2020) . |
[24] | Chung A. Figure 1 COVID-19 chest x-ray data initiative. https://github.com/agchung/Figure1-COVID-chestxray-dataset. (2020) . |
[25] | Pattnaik G, Shrivastava VK, Parvathi K. Transfer Learning-Based Framework for Classification of Pest in Tomato Plants. Appl Artif Intell. Taylor & Francis. (2020) ; 34: (13): 981–93. |
[26] | Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L. Imagenet: A large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition. IEEE. (2009) ; pp. 248–55. |
[27] | Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv Prepr arXiv14091556. (2014) . |
[28] | He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. (2016) ; pp. 770–8. |
[29] | Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. (2015) ; pp. 1–9. |
[30] | Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXivPrepr arXiv150203167. (2015) . |
[31] | Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. (2016) ; pp. 2818–26. |
[32] | Szegedy C, Ioffe S, Vanhoucke V, Alemi AA. Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-First AAAI Conference on Artificial Intelligence. (2017) . |
[33] | Chollet F. Xception: Deep learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. (2017) ; pp. 1251–8. |
[34] | Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, et al. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv Prepr arXiv170404861. (2017) . |
[35] | Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC. Mobilenetv2: Inverted residuals and linear bottlenecks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. (2018) ; pp. 4510–20. |
[36] | Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. (2017) ; pp. 4700–8. |
[37] | Alzammam A, Binsalleeh H, AsSadhan B, Kyriakopoulos KG, Lambotharan S. Comparative analysis on imbalanced multi-class classification for malware samples using CNN. In: 2019 International Conference on Advances in the Emerging Computing Technologies (AECT). IEEE. (2020) ; pp. 1–6. |
[38] | Elasnaoui K, Chawki Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J Biomol Struct Dyn. Taylor & Francis. (2020) ; 1–22. |




